Abstract
In the last 30 years, twin studies have indicated a strong genetic contribution to Autism Spectrum Disorders (ASD). The heritability of ASD is estimated to be 50 %, mostly captured by still unknown common variants. In approximately 10 % of patients with ASD, especially those with intellectual disability, de novo copy number or single nucleotide variants affecting clinically relevant genes for ASD can be identified. Given the function of these genes, it was hypothesized that abnormal synaptic plasticity and failure of neuronal/synaptic homeostasis could increase the risk of ASD. In parallel, abnormal levels of blood serotonin and melatonin were reported in a subset of patients with ASD. These biochemical imbalances could act as risk factors for the sleep/circadian disorders that are often observed in individuals with ASD. Here, we review the main pathways associated with ASD, with a focus on the roles of the synapse and the serotonin-NAS-melatonin pathway in the susceptibility of ASD.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Autism Spectrum Disorder
- Autism Spectrum Disorder
- Intellectual Disability
- Intellectual Disability
- Social Responsiveness Scale
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Autism Spectrum Disorders (ASD) are a group of neuropsychiatric disorders characterized by problems in social communication as well as the presence of restricted interests and stereotyped and repetitive behaviors (Kanner 1943; Asperger 1944; Coleman and Gillberg 2012). Epidemiological studies estimate that more than 1 % of the population could receive a diagnosis of ASD (Elsabbagh et al. 2012; Developmental Disabilities Monitoring Network Surveillance Year Principal 2014). Individuals with ASD can also suffer from other psychiatric and medical conditions, including intellectual disability (ID), epilepsy, motor control difficulties, Attention-Deficit Hyperactivity Disorder (ADHD), tics, anxiety, sleep disorders, epilepsy, depression or gastrointestinal problems (Gillberg 2010; Moreno-De-Luca et al. 2013). The term ESSENCE, for ‘Early Symptomatic Syndromes Eliciting Neurodevelopmental Clinical Examinations,’ was coined by Christopher Gillberg to take into account this clinical heterogeneity and syndrome overlap (Gillberg 2010). There are four to eight times more males than females with ASD (Elsabbagh et al. 2012), but the sex ratio is more balanced in patients with ID and/or dysmorphic features (Miles et al. 2005). Autism can be studied as a category (affected vs. unaffected) or as a quantitative trait using auto- or hetero-questionnaires such as the Social Responsiveness Scale (SRS) or the autism quotient (AQ) (Ronald et al. 2006; Skuse et al. 2009; Constantino 2011). Using these tools, autistic traits seem to be normally distributed in clinical cases as well as in the general population (Ronald et al. 2006; Skuse et al. 2009; Constantino 2011).
The causes of autism remain largely unknown, but twin studies have constantly shown a high genetic contribution to ASD. Molecular genetics studies have identified more than 100 ASD risk genes carrying rare and penetrant deleterious mutations in approximately 10–25 % of patients (Huguet et al. 2013; Gaugler et al. 2014; Bourgeron 2015). In addition, quantitative genetics studies have shown that common genetic variants could capture almost all the heritability of ASD (Huguet et al. 2013; Gaugler et al. 2014). The genetic landscape of ASD is shaped by a complex interplay between common and rare variants and is most likely different from one individual to another (Gardener et al. 2011; Hallmayer et al. 2011; Bourgeron 2015). Remarkably, the susceptibility genes seem (Huguet et al. 2013) to converge in a limited number of biological pathways, including chromatin remodeling, protein translation, actin dynamics and synaptic functions (Bourgeron 2009; Toro et al. 2010; Huguet et al. 2013; Bourgeron 2015). In addition, several studies have pointed to a dysfunction of the serotonin-NAS-melatonin pathway in patients with ASD. Abnormalities of this pathway might increase the risk of circadian/sleep disorders often observed in patients with ASD.
In this chapter, we will detail the advances in the genetics of ASD (Fig. 1) with a focus on the role of both synapses and biological rhythms in the susceptibility of ASD (Abrahams and Geschwind 2008; Bourgeron 2009; Toro et al. 2010; Devlin and Scherer 2012; Huguet et al. 2013).
The history of the genetics of autism from 1975 to 2015. The increase in the identified genes associated with ASD (SFARI—March 2015) is represented together with the prevalence of ASD (data taken from the Center for Disease Control and Prevention), the different versions of the Diagnostic Statistical Manual (from DSM II to DSM 5.0) and the advance in genetics technology (Adapted from Huguet and Bourgeron 2016)
Twin and Family Studies in ASD
Based on more than 13 twin studies published between 1977 and 2015, researchers have estimated the genetic and environmental contribution to ASD (Fig. 2). In 1977, the first twin study of autism by Folstein and Rutter (1977) reported on a cohort of 11 monozygotic (MZ) twins and 10 dizygotic (DZ) twins. This study showed that MZ twins were more concordant for autism—36 % (4/11)—compared with 0 % (0/10) for DZ twins. When a “broader autism phenotype” was used, the concordance increased to 92 % for MZ twins and to 10 % DZ twins (Bailey et al. 1995). Since this first small scale study, twin studies have constantly reported a higher concordance for ASD in MZ compared with DZ (Ritvo et al. 1985; Steffenburg et al. 1989; Bailey et al. 1995; Le Couteur et al. 1996). Between 2005 and 2009, three twin studies with relatively large groups of twins (285–3419) have reported high concordances for ASD in MZ twins (77–95 %) compared with DZ twins (31 %; Ronald et al. 2005; Taniai et al. 2008; Rosenberg et al. 2009). Notably, MZ concordances were similar to those reported in the previous studies, but DZ concordances were higher. In 2010, Lichtenstein et al. reported a relatively low concordance for ASD in 39 % of the MZ twins compared with other studies (the concordance for DZ twins in this study was 15 %). However, as previously indicated by studies using the “broader autism phenotype,” all discordant MZ twins of this cohort had symptoms of ESSENCE (e.g., ID, ADHD, language delay, etc.). A significant proportion of the genetic contribution to ASD was shown to be shared with other neurodevelopmental disorders such as ADHD (>50 %) and learning disability (>40 %; Lichtenstein et al. 2010; Ronald et al. 2010; Lundstrom et al. 2011; Ronald and Hoekstra 2011). In summary, when all twin studies are taken into account, concordance for ASD is roughly 45 % for MZ twins and 16 % for DZ twins (Ritvo et al. 1985; Steffenburg et al. 1989; Bailey et al. 1995; Le Couteur et al. 1996).
The main twins studies in ASD. A total of 13 twins studies and 17 heritability studies are depicted. Means of concordance and heritability weighted by sample size are presented on the right of the figure (Adapted from Huguet and Bourgeron 2016)
Family studies also showed that the recurrence of having a child with ASD increases with the proportion of the genome that the individual shares with one affected sibling or parent (Constantino et al. 2010; Risch et al. 2014; Sandin et al. 2014). In a population-based sample of 14,516 children diagnosed with ASD (Sandin et al. 2014), the relative risk for ASD (compared to the general population) was estimated to be 153.0 [95 % confidence interval (CI): 56.7–412.8] for MZ twins, 8.2 (3.7–18.1) for DZ twins; 10.3 (9.4–11.3) for full siblings, 3.3 (95 % CI, 2.6–4.2) for maternal half siblings, 2.9 (95 % CI, 2.2–3.7) for paternal half siblings, and 2.0 (95 % CI, 1.8–2.2) for cousins.
Heritability is the proportion of the phenotypic variation in a trait of interest, measured in a given studied population and in a given environment, that is co-varying with genetic differences among individuals in the same population. In 1995, based on a twin study, Bailey et al. estimated the heritability of autism to be 91–93 %. Since then, the estimation of heritability has differed from one study to another, but the genetic variance has accounted for at least 38 % and up to 90 % of the phenotypic variance (Hallmayer et al. 2011; Ronald and Hoekstra 2011; Sandin et al. 2014). Using a large cohort of 14,516 children diagnosed with ASD Sandin et al. (2014), estimated the heritability to be 0.50 (95 % CI, 0.45–0.56) and the non-shared environmental influence was also 0.50 (95 % CI, 0.44–0.55). Surprisingly, only the additive genetic component and the non-shared environment seemed to account for the risk of developing ASD (Sandin et al. 2014).
In summary, epidemiological studies provide crucial information about the heritability of ASD. However, they do not inform us about the genes involved or the number and frequency of their variants. In the last 15 years, candidate genes and whole-genome analyses have been performed to address these questions.
From Chromosomal Rearrangements to Copy Number Variants in ASD
The first genetic studies that associated genetic variants with ASD used observations from cytogenetic studies (Gillberg and Wahlstrom 1985). However, because of the low resolution of the karyotypes (several Mb), it was almost impossible to associate a specific gene to ASD using this approach. The prevalence of large chromosomal abnormalities is estimated to be less than 2 % (Vorstman et al. 2006). Thanks to progress in molecular technologies such as Comparative Genomic Hybridization (CGH) or SNP arrays, the resolution in the detection of genomic imbalances has dramatically increased. Depending on the platforms, Copy Number Variants (CNVs) of more than 50 kb are now robustly detected (Pinto et al. 2011). Since the first articles published in 2006, a very large number of studies have investigated the contribution of CNVs to ASD (Jacquemont et al. 2006; Sebat et al. 2007). Several studies using the Simons Simplex Collection could even provide an estimation of the frequency of the de novo CNVs in patients with ASD compared with their unaffected siblings (Sanders et al. 2011). All together, de novo CNVs are present in 4–7 % of the patients with ASD compared to 1–2 % in the unaffected siblings and controls (Glessner et al. 2009; Sanders et al. 2011; Pinto et al. 2014). The studies have also indicated that de novo CNVs identified in patients are most likely altering genes and most especially genes associated with synaptic functions and/or regulated by FMRP, the protein responsible for the fragile X syndrome (Pinto et al. 2010, 2014). Beyond ASD, large CNVs (>400 kb) affecting exons are present in 15 % of patients with Developmental Delay (DD) or ID (Cooper et al. 2011). Most of the CNVs are private to each individual, but some are recurrently observed in independent patients. For example, three loci on chromosomal regions 7q11, 15q11.2–13.3, and 16p11.2 have been strongly associated with ASD (Ballif et al. 2007; Kumar et al. 2008; Weiss et al. 2008; Szafranski et al. 2010; Sanders et al. 2011; Leblond et al. 2014).
In summary, large chromosomal rearrangements and CNVs increase the risk of having ASD in 5–10 % of the individuals (Vorstman et al. 2006; Pinto et al. 2010, 2014). To go further in the identification of the ASD risk genes, candidate genes and whole exome/genomes studies were performed.
From Candidate Genes to Whole Exome/Genome Sequencing Studies in ASD
The first approach to associate a gene with ASD was to select specific candidate genes based on data coming from functional or genetic studies or a combination of the two. This approach was successful in identifying several synaptic genes associated with ASD such as NLGN3, NLGN4X, SHANK3 and NRXN1 (Jamain et al. 2003; Durand et al. 2007; Szatmari et al. 2007). Thanks to the advance in Next Generation Sequencing (NGS), we can now interrogate all genes of the genome in an unbiased manner using Whole Exome/Genome Sequencing (WES, WGS).
To date, more than 18 WES studies of sporadic cases of ASD (O’Roak et al. 2011, 2012a; Chahrour et al. 2012; Iossifov et al. 2012; Neale et al. 2012; Sanders et al. 2012; He et al. 2013; Lim et al. 2013; Liu et al. 2013, 2014; Willsey et al. 2013; Yu et al. 2013; An et al. 2014; De Rubeis et al. 2014; Iossifov et al. 2014; Samocha et al. 2014; Chang et al. 2015; Krumm et al. 2015) have been performed, comprising altogether more than >4000 families (Table 1). In almost all these studies, the authors have especially focused their analysis on the contribution of de novo Single Nucleotide Variants (SNVs) in ASD. All together, the average number of de novo coding SNVs per individual (including missense, splicing, frameshift, and stop-gain variants) is estimated to be approximately 0.86 in female patients, 0.73 in male patients, and 0.60 in unaffected male and female siblings (Krumm et al. 2014; Ronemus et al. 2014). Interestingly, de novo SNVs were three times more likely to be on the paternal chromosome than on the maternal one (Kong et al. 2012; O’Roak et al. 2012a) with an increase of almost two de novo mutations per year and doubled every 16.5 years (Kong et al. 2012).
Based on these studies (Iossifov et al. 2012; Neale et al. 2012; O’Roak et al. 2012a; Sanders et al. 2012), 3.6–8.8 % of the patients were shown to carry a de novo causative mutation (Iossifov et al. 2012) with a twofold increase of deleterious mutations in the patients compared with their unaffected siblings. In a meta-analysis, using more than 2500 families, Iossifov et al. (2014) found that de novo Likely Gene Disrupting (LGD) mutations (frameshift, nonsense and splice site) were more frequent in patients with ASD compared with unaffected siblings (P = 5 × 10−7). The carriers of these de novo LGDs were more likely diagnosed with a low non-verbal IQ. The de novo LGDs are significantly enriched in genes involved in chromatin modeling factors (P = 4 × 10−6) and in genes regulated by the FMRP complex (p = 4 × 10−7). Following these whole exome studies, targeted re-sequencing studies of the most compelling candidate genes were performed (O’Roak et al. 2012b). All together, 10 genes carrying de novo mutations were significantly associated with ASD: CHD8, DYRK1A, GRIN2B, KATNAL2, RIMS1, SCN2A, POGZ, ADNP, ARID1B and TBR1.
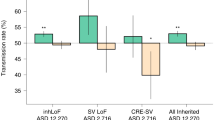
Only a few studies have analyzed the contribution of inherited SNVs in ASD. In 2013, Lim et al. analyzed whole exome sequencing of 933 cases (ASD) and 869 controls for the presence of rare complete human knockouts (KO) with homozygous or compound heterozygous loss-of-function (LoF) variants (≤5 % frequency). They observed a significant twofold increase in complete KOs in patients with ASD compared to controls. They estimated that such complete KO mutations could account for 3 % of the patients with ASD. For the X chromosome, there was a significant 1.5-fold increase in complete KO in affected males compared to unaffected males that could account for 2 % of males with ASD (Lim et al. 2013). The same year, Yu et al. (2013) analyzed 104 consanguineous families including 79 families with a single child with ASD (simplex families) and 25 families with more than one affected individual (multiplex families) collected by the Homozygosity Mapping Collaborative for Autism (HMCA). They identified biallelic mutations in AMT, PEX7, SYNE1, VPS13B, PAH, and POMGNT1. Finally, a very recent study by Krumm et al. (2015) ascertained the relative impact of inherited and de novo variants (CNVs or SNVs) on ASD risk in 2377 families. Inherited truncating variants were enriched in probands (for SNV odds ratio = 1.14, P = 0.0002; for CNV odds ratio = 1.23, P = 0.001) in comparison to unaffected siblings (Krumm et al. 2015). Interestingly, they also observed a significant maternal transmission bias of inherited LGD to sons. New ASD-risk genes were also identified such as RIMS1, CUL7 and LZTR1.
To date, few whole genome sequencing studies have been published for ASD (Table 1). Michaelson et al. (2012) analyzed 40 WGS of monozygotic twins concordant for ASD and their parents. They proposed that ASD-risk genes could be hot spots of mutation in the genome and confirmed the association between ASD and de novo mutations in GPR98, KIRREL3 and TCF4. Shi et al. (2013) analyzed a large pedigree with two sons affected with ASD and six unaffected siblings, focusing on inherited mutations. They identified ANK3 as the most likely candidate gene. In 2015, Yuen et al. analyzed 85 families with two children affected with ASD. This study represents the largest published WGS data set in ASD. They identified 46 ASD-relevant mutations present in 36 of 85 (42.4 %) families. Only 16 ASD-relevant mutations of 46 (35 %) identified were de novo. Very interestingly, for more than half of the families (69.4 %; 25 of 36), the two affected siblings did not share the same rare penetrant ASD risk variant(s).
Whole genome sequencing is also very efficient to identify mutations in regions of the human genome that are wrongly annotated and in regions that are highly GC rich. For example, mutations on the SHANK3 gene were rarely identified using whole exome sequencing, given its high GC content (Leblond et al. 2014). In contrast, whole genome sequencing could successfully identify SHANK3 mutations (Nemirovsky et al. 2015; Yuen et al. 2015).
The Common Variants in ASD
In the general population, one individual carries on average three million genetic variants in comparison to the reference human genome sequence (Xue et al. 2012; Fu et al. 2013; Genome of the Netherlands and Genome of the Netherlands 2014). The vast majority of the variants (>95 %) are the so-called common variants shared with more than 5 % of the human population (Xue et al. 2012; Fu et al. 2013; Genome of the Netherlands and Genome of the Netherlands 2014). While there is not a clear border between common and rare variants, it is nevertheless interesting to estimate the role of the genetic variants found in the general population in the susceptibility to ASD.
Using quantitative genetics, Klei et al. (2012) estimated that common variants were contributing to a high proportion of the liability of ASD: 40 % in simplex families and 60 % in multiplex families. In 2014, a study by Gaugler et al. used the same methodology (Yang et al. 2011) and provided an estimation of the heritability (52.4 %) that is almost exclusively due to common variation, leaving only 2.6 % of the liability to the rare variants. The contribution of common variants is therefore important, but unfortunately the causative SNPs still remain unknown since they are numerous (>1000) and each is associated with a low risk. To date, the largest genome wide association studies (GWAS) performed on <5000 families with ASD were underpowered to identify a single SNP with genome wide significance (Anney et al. 2012; Cross-Disorder Group of the Psychiatric Genomics 2013).
The recruitment of larger cohorts of patients with dimensional phenotypes is therefore warranted to better ascertain the heritability of ASD and to identify the genetic variants, which explain most of the genetic variance.
The Genetic Architecture of ASD
Based on the results obtained from epidemiological and molecular studies, it is now accepted that the genetic susceptibility to ASD can be different from one individual to another with a combination of rare deleterious variants (R) and a myriad of low-risk alleles [also defined as the genetic background (B)]. Most of the inherited part of ASD seems to be due to common variants observed in the general population, with only a small contribution from rare variants (Fig. 3). Importantly, while the de novo mutations are considered per se as genetic factors, they do not contribute to the heritability since they are only present in the patient (with the relatively rare exception of germinal mosaicisms present in one of the parental germlines and transmitted to multiple children). These de novo events could therefore be considered as “environmental causes” of ASD but acting on the DNA molecule. It is currently estimated that more than 500–1000 genes could account for these “monogenic forms” of ASD (Iossifov et al. 2012; Sanders et al. 2012), confirming the high degree of genetic heterogeneity.
Relative contribution of the genetics and environment in ASD. Based on twin and familial studies, it is estimated that the genetic and environmental contributions to ASD are approximately 50/50 %. Most of the heritable part seems to be due to common variants observed in the general population, with a small contribution of rare variants. Importantly, the de novo mutations are genetic causes of ASD but do not contribute to the heritability since there are only present in the patient. These de novo events are therefore considered to be “environmental causes” of ASD, but acting on the DNA molecule (Adapted from Huguet and Bourgeron 2016)
The interplay between the rare or de novo variants R and the background B will also influence the phenotypic diversity observed in the patients carrying rare deleterious mutations. In some individuals, a genetic background B will be able to buffer or compensate the impact of the rare genetic variations R. In contrast, in some individuals, the buffering capacity of B will not be sufficient to compensate the impact of R and they will develop ASD (Rutherford 2000; Hartman et al. 2001). In the R‘n’B model, ASD can be regarded as a collection of many genetic forms of “autisms,” each with a different etiology ranging from monogenic to polygenic models.
The presence of multiple hits of rare CNVs, SNVs or indels in a single individual also illustrates the complexity of the genetic landscape of ASD (Girirajan et al. 2010, 2012; Leblond et al. 2012). In addition, the analysis of the whole genome sequence of multiplex families also indicates that clinically relevant mutations can be different from one affected sib to another even in a single family (Yuen et al. 2015). It is therefore still difficult to ascertain robust genotype-phenotype relationships based on our current knowledge.
Fortunately, although the ASD-risk genes are numerous, they seem to converge in a limited number of biological pathways that are currently scrutinized by many researchers.
The Biological Pathways Associated with ASD
Unbiased pathway analyses indicated that ASD-risk genes seem to be enriched in groups of proteins with specific functions (Voineagu et al. 2011; De Rubeis et al. 2014; Ronemus et al. 2014; Uddin et al. 2014; Hormozdiari et al. 2015). Pinto et al. (2014) analyzed the burden of CNVs in 2446 individuals with ASD and 2640 controls and found enrichment in genes coding post-synaptic density proteins and FMRP targets. Ronemus et al. (2014) reviewed the results of four whole exome sequencing studies and showed an enrichment of mutated genes in chromatin modifier genes (P = 4 × 10−6) and FMRP targets (P = 7 × 10−6). Protein-protein interactions (PPI) analyses of the genes carrying LGD mutations also showed enrichment in proteins involved in neuronal development and axon guidance, signaling pathways and chromatin and transcription regulation. De Rubeis et al. (2014) also used the PPI network and showed enrichment in clusters of proteins involved in the cell junction TGF beta pathway, cell communication and synaptic transmission, neurodegeneration and transcriptional regulation.
In parallel to the genetic studies, several transcriptomic analyses were performed using post-mortem brain of individuals with ASD (Voineagu et al. 2011; Gupta et al. 2014). Several genes were differentially expressed or correlated between brain regions. Two network modules were identified. The first module was related to interneurons and to genes involved in synaptic function (downregulated in brains from ASD patients compared to controls). The second module was related to immunity and microglia activation (upregulated in brains from ASD patients compared to controls).
Based on these results, neurobiological studies using cellular and animal models have been performed to identify the main mechanisms leading to ASD. Remarkably, several studies showed that neuronal activity seems to regulate the function of many of the ASD-risk genes, leading to the hypothesis that abnormal synaptic plasticity and failure of neuronal/synaptic homeostasis could play a key role in the susceptibility to ASD (Belmonte and Bourgeron 2006; Auerbach et al. 2011; Toro et al. 2010). Here, we will only depict four main biological pathways associated with ASD: chromatin remodeling, protein synthesis, protein degradation, and synaptic function (Fig. 4). In parallel, several biochemical studies have indicated a dysfunction in the serotonin-NAS-melatonin pathway.
Examples of the biological pathways associated with ASD. The ASD-risk genes code for proteins involved in chromatin remodeling, transcription, protein synthesis and degradation, cytoskeleton dynamics, and synaptic functions. Proteins associated with ASD are circled in red (Adapted from Huguet and Bourgeron 2016)
Chromatin Remodeling
Mutations in genes encoding key regulators of chromatin remodeling and gene transcription (e.g., MECP2, MEF2C, HDAC4, CHD8 and CTNNB1) have been reported in individuals with ASD (Fig. 4). Remarkably, a subset of these genes is regulated by neuronal activity and influences neuronal connectivity and synaptic plasticity (Cohen et al. 2011; Sando et al. 2012; Ebert et al. 2013).
Protein Synthesis
The level of synaptic proteins can be influenced by neuronal activity through global and local synaptic mRNA translation (Ma and Blenis 2009). Several genes involved in such activity-driven regulation of synaptic proteins have been found to be mutated in individuals with ASD (Kelleher and Bear 2008). For example, the mTOR pathway controls global mRNA translation and its deregulation causes diseases associated with increased cell proliferation and loss of autophagy, including cancer (Ma and Blenis 2009), but also increases the risk for ASD. Remarkably, mutations in the repressor of the mTOR pathway such as NF1, PTEN and SynGAP1 cause an increase of translation in neurons and at the synapse (Auerbach et al. 2011). Mutations of the FMRP–EIF4E–CYFIP1 complex cause the fragile X syndrome and increase the risk of ASD (Budimirovic and Kaufmann 2011). This protein complex controls local translation of mRNA at the synapse and acts downstream of the Ras-ERK signaling pathway. This complex regulates the translation of more than 1000 specific genes, many of which are ASD risk genes (De Rubeis et al. 2013; Fernandez et al. 2013; Gkogkas et al. 2013; Santini et al. 2013). An alteration of this FMRP–EIF4E–CYFIP1 complex should therefore create an imbalance in the level of many synaptic proteins that are associated with ASD.
Protein Degradation
The Ubiquitin-Proteasome System (UPS) is central for the degradation of the proteins and, consequently, for the regulation of synapse composition, assembly and elimination (Mabb and Ehlers 2010). The UBE3A gene encodes a ubiquitin ligase, is mutated in patients with Angelman syndrome and is duplicated on the maternal chromosome 15q11 in individuals with ASD. Neuronal activity increases UBE3A transcription through the MEF2 complex and regulates excitatory synapse development by controlling the degradation of ARC, a synaptic protein that decreases long-term potentiation by promoting the internalization of AMPA receptors (Greer et al. 2010).
Synaptic Functions
Many proteins encoded by ASD-risk genes participate in different aspects of neuronal connectivity, such as glutamatergic (e.g., GRIN2B), GABAergic (e.g., GABRA3 and GABRB3) and glycinergic (e.g., GLRA2) neurotransmission, neuritogenesis (e.g., CNTN), the establishment of synaptic identity (e.g., cadherins and protocadherins), neuronal conduction (CNTNAP2) and permeability to ions (CACNA1, CACNA2D3 and SCN1A). Some of these proteins are directly involved in activity-driven synapse formation, such as the neurexins (NRXNs) and neuroligins (NLGNs). Some are scaffolding proteins involved in the positioning of cell-adhesion molecules and neurotransmitter receptors at the synapse (Sheng and Kim 2011; Choquet and Triller 2013). For example, deletions, duplications and coding mutations in the three SHANK genes (SHANK1, SHANK2 and SHANK3) have been recurrently reported in individuals with ASD (Leblond et al. 2014). SHANK proteins assemble into large molecular platforms in interaction with glutamate receptors and actin-associated proteins (Grabrucker et al. 2011). In vitro, SHANK3 mutations identified in individuals with ASD reduce actin accumulation in spines affecting the development and morphology of dendrites as well as the axonal growth cone motility (Durand et al. 2012). In mice, mutations in SHANK3 decrease spine density in the hippocampus but also increase dendritic arborizations in striatal neurons (Peca et al. 2011). Mice mutated in SHANK present with behavior resembling autistic features in humans. Shank1 knockout mice display increased anxiety, decreased vocal communication, decreased locomotion and, remarkably, enhanced working memory, but decreased long-term memory (Hung et al. 2008; Silverman et al. 2011; Wohr et al. 2011). Shank2 knockout mice present hyperactivity, increased anxiety, repetitive grooming, and abnormalities in vocal and social behaviors (Schmeisser et al. 2012; Won et al. 2012). Shank3 knockout mice show self-injurious repetitive grooming, and deficits in social interaction and communication (Bozdagi et al. 2010; Peca et al. 2011; Wang et al. 2011; Yang et al. 2012).
The Serotonin-NAS-Melatonin Pathway
In parallel to the genetic investigations pointing to the biological pathways described above, several biological abnormalities have been reported in individuals with ASD, including neurochemical, immunological, endocrine or metabolic traits (Lam et al. 2006; Rossignol and Frye 2012), which may provide insights into the pathophysiology of ASD. Among these, elevated blood serotonin is one of the most replicated findings (Pagan et al. 2012) (Fig. 5). A deficit in melatonin (which chemically derives from serotonin) in the blood or urine of individuals with ASD has also been described in several studies (Tordjman et al. 2005; Melke et al. 2008) and is associated with increased peripheral N-acetylserotonin (NAS), the intermediate metabolite between serotonin and melatonin. Several mutations affecting this pathway were identified but the mechanisms of these biochemical impairments remain mostly unexplained. Melatonin is a neurohormone mainly synthesized in the pineal gland during the night. It is a biological signal of light/dark cycles and is considered to be a major circadian synchronizer. It is also a modulator of metabolism, immunity, reproduction and digestive function. Furthermore, it displays antioxidant and neuroprotective properties and can directly modulate neuronal networks (Bourgeron 2007). Melatonin appears as a therapeutic target of the frequently reported sleep disorders associated with ASD (Andersen et al. 2008; Wright et al. 2011; Malow et al. 2012). NAS displays intrinsic biological properties: it is an agonist of the TrkB receptor and may thus share the neurotrophic properties of brain-derived neurotrophic factor (BDNF), the canonical TrkB ligand (Jang et al. 2010; Sompol et al. 2011). Serotonin conversion into melatonin involves two sequential enzymatic steps: N-acetylation of serotonin into NAS by arylalkylamine N-acetyltranferase (AANAT, EC: 2.3.1.87) followed by methylation by acetylserotonin O-methyltransferase (ASMT, also called hydroxyindole O-methyltransferase HIOMT, EC: 2.1.1.4) (Fig. 5a). We previously showed that deleterious mutations of the ASMT gene could disrupt melatonin synthesis in a subset of patients with ASD. Nevertheless, the frequency of such a deleterious mutation is too low (2 % of the cases) to explain the relatively high frequency of melatonin deficit in ASD (>50 % of the patients, taking as a threshold the fifth percentile of the controls). More recently, we observed a low level of the 14-3-3 proteins both in the blood platelet and pineal gland of patients with ASD (Pagan et al. 2014). These ubiquitous chaperone proteins are known to form a protein complex, the ‘melatoninosome,’ involving AANAT and ASMT in pinealocytes (Obsil et al. 2001; Maronde et al. 2011). This interaction between 14-3-3 and AANAT and/or ASMT might be essential for the production of melatonin and an adequate balance of the serotonin-NAS-melatonin pathway. Indeed, a low level of 14-3-3 could eventually lead to a deficit in enzyme activity, contributing to the global disruption of the serotonin-NAS-melatonin pathway observed in ASD. Studies investigating the regulation of the complex 14-3-3/ASMT/AANAT in ASD and controls are in progress.
The serotonin-NAS-melatonin pathway in ASD. (a) The serotonin-NAS-melatonin synthesis pathway consists of two enzymatic steps involving first the N-acetylation of serotonin to N-acetylserotonin (NAS) by the rate-limiting enzyme AANAT and the methylation of NAS by the ASMT (also known as HIOMT). Different alterations such as higher serotonin or low melatonin levels were observed in the blood of patients with ASD. The enzymes are represented in gray and metabolites are in white. Alterations of the biochemical parameters are shown with red arrows. (b) A schematic view of the pineal gland with the pinealocytes that contain the “melatoninosome.” This complex includes at least four proteins: AANAT, ASMT, 14-3-3 and S-antigen. The immunofluorescence confocal image of AANAT (green) and 14-3-3δ/ε (red) in the pinealocytes is adapted from Maronde et al. (2011). The structure of the 14-3-3ζ homodimer binding to AANAT is adapted from Obsil et al. (2001). (c) Main sources of serotonin (blue) and melatonin (green) and the symptoms or comorbidities of ASD relevant to alterations in serotonin and melatonin levels observed in ASD (Adapted from Pagan et al. 2012)
Perspectives
In the last 30 years, very significant progress has been made in the genetics of ASD. We now have a better knowledge on the genetic architecture of this heterogeneous syndrome and some of the biological pathways have been investigated using different approaches such as cellular and animal models. There are, however, many aspects of the genetics of ASD that remain largely unknown.
The first challenge concerns the role of the common variants. These variants are most likely playing a key role in the susceptibility to ASD and in the severity of the symptoms. But, because the impact of each single SNP is very low, it is currently impossible to identify the risk alleles using conventional GWAS. In human quantitative traits such as height, neuroanatomical diversity or intellectual quotient, very large cohorts of many thousands of individuals are necessary to identify the main causative SNPs (Toro et al. 2014; Yang et al. 2010; Deary et al. 2012).
The second challenge concerns the stratification of the patients and the role of the ASD-risk genes during brain development/function. Based on our current knowledge, the genetic architecture of ASD seems to be different from one individual to another, with possibly contrasting impact on when and where neuronal connectivity could be atypical compared to the general population. For example, in animal models, several mutations lead to higher connectivity whereas other mutations alter synaptic density. It is therefore crucial to increase our knowledge from a basic research perspective about the biological roles of the ASD-risk genes and their partners.
Finally, while we all agree that biological research is necessary to improve the quality of life of the patients and their families (for example, to alleviate the comorbidities associated with ASD like sleep and gastrointestinal problems), progress should also be made toward better recognition and inclusion of people with neuropsychiatric conditions in our societies (no mind left behind). Hopefully, increasing knowledge in genetics, neurology and psychology should allow for better diagnosis, care for and integration of individuals with autism.
References
Abrahams BS, Geschwind DH (2008) Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 9:341–355
An JY, Cristino AS, Zhao Q, Edson J, Williams SM, Ravine D, Wray J, Marshall VM, Hunt A, Whitehouse AJ, Claudianos C (2014) Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach. Transl Psychiatry 4:e394
Andersen IM, Kaczmarska J, McGrew SG, Malow BA (2008) Melatonin for insomnia in children with autism spectrum disorders. J Child Neurol 23:482–485
Anney R et al (2012) Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet 21:4781–4792
Asperger H (1944) Die “autistischen Psychopathen” im Kindesalter. Arch Psychiatr Nervenkr 177:76–137
Auerbach BD, Osterweil EK, Bear MF (2011) Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature 480:63–68
Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77
Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, Graham JM Jr, Medne L, Zackai EH, Shaikh TH, Geoghegan J, Selzer RR, Eis PS, Bejjani BA, Shaffer LG (2007) Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet 39:1071–1073
Belmonte MK, Bourgeron T (2006) Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci 9:1221–1225
Bourgeron T (2007) The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harb Symp Quant Biol 72:645–654
Bourgeron T (2009) A synaptic trek to autism. Curr Opin Neurobiol 19:231–234
Bourgeron T (2015) From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci 16:551–563
Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD (2010) Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism 1:15
Budimirovic DB, Kaufmann WE (2011) What can we learn about autism from studying fragile X syndrome? Dev Neurosci 33:379–394
Chahrour MH, Yu TW, Lim ET, Ataman B, Coulter ME, Hill RS, Stevens CR, Schubert CR, Collaboration AAS, Greenberg ME, Gabriel SB, Walsh CA (2012) Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet 8:e1002635
Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D (2015) Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci 18:191–198
Choquet D, Triller A (2013) The dynamic synapse. Neuron 80:691–703
Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME (2011) Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72:72–85
Coleman M, Gillberg C (2012) The autisms. Oxford University Press, New York
Constantino JN (2011) The quantitative nature of autistic social impairment. Pediatr Res 69:55R–62R
Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P (2010) Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry 167:1349–1356
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE (2011) A copy number variation morbidity map of developmental delay. Nat Genet 43:838–846
Cross-Disorder Group of the Psychiatric Genomics C (2013) Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381:1371–1379
De Rubeis S, Pasciuto E, Li KW, Fernandez E, Di Marino D, Buzzi A, Ostroff LE, Klann E, Zwartkruis FJ, Komiyama NH, Grant SG, Poujol C, Choquet D, Achsel T, Posthuma D, Smit AB, Bagni C (2013) CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron 79:1169–1182
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimäki T, Lin CF, Ma'ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, DDD Study, Homozygosity Mapping Collaborative for Autism, UK10K Consortium, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515:209–215
Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, Luciano M, Lopez LM, Gow AJ, Corley J, Redmond P, Fox HC, Rowe SJ, Haggarty P, McNeill G, Goddard ME, Porteous DJ, Whalley LJ, Starr JM, Visscher PM (2012) Genetic contributions to stability and change in intelligence from childhood to old age. Nature 482:212–215
Developmental Disabilities Monitoring Network Surveillance Year Principal I (2014) Prevalence of autism spectrum disorder among children aged 8 years – autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ 63(Suppl 2):1–21
Devlin B, Scherer SW (2012) Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev 22:229–237
Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T (2007) Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 39:25–27
Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N (2012) SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry 17:71–84
Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, Greenberg ME (2013) Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499:341–345
Elsabbagh M, Divan G, Koh YJ, Kim YS, Kauchali S, Marcin C, Montiel-Nava C, Patel V, Paula CS, Wang C, Yasamy MT, Fombonne E (2012) Global prevalence of autism and other pervasive developmental disorders. Autism Res 5:160–179
Fernandez E, Rajan N, Bagni C (2013) The FMRP regulon: from targets to disease convergence. Front Neurosci 7:191
Folstein S, Rutter M (1977) Genetic influences and infantile autism. Nature 265:726–728
Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Project NES, Akey JM (2013) Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–220
Gardener H, Spiegelman D, Buka SL (2011) Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis. Pediatrics 128:344–355
Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, Ripke S, Sandin S, Sklar P, Svantesson O, Reichenberg A, Hultman CM, Devlin B, Roeder K, Buxbaum JD (2014) Most genetic risk for autism resides with common variation. Nat Genet 46:881–885
Genome of the Netherlands C, Genome of the Netherlands C (2014) Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46:818–825
Gillberg C (2010) The ESSENCE in child psychiatry: early symptomatic syndromes eliciting neurodevelopmental clinical examinations. Res Dev Disabil 31:1543–1551
Gillberg C, Wahlstrom J (1985) Chromosome abnormalities in infantile autism and other childhood psychoses: a population study of 66 cases. Dev Med Child Neurol 27:293–304
Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE (2010) A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 42:203–209
Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, Nezarati MM, Asamoah A, Jackson KE, Gowans GC, Martin JA, Carmany EP, Stockton DW, Schnur RE, Penney LS, Martin DM, Raskin S, Leppig K, Thiese H, Smith R, Aberg E, Niyazov DM, Escobar LF, El-Khechen D, Johnson KD, Lebel RR, Siefkas K, Ball S, Shur N, McGuire M, Brasington CK, Spence JE, Martin LS, Clericuzio C, Ballif BC, Shaffer LG, Eichler EE (2012) Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 367:1321–1331
Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille JC, Sonenberg N (2013) Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493:371–377
Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459:569–573
Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM (2011) Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol 21:594–603
Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME (2010) The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140:704–716
Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE (2014) Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun 5:5748
Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N (2011) Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry 68:1095–1102
Hartman JL, Garvik B, Hartwell L (2001) Principles for the buffering of genetic variation. Science 291:1001–1004
He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, Schellenberg GD, Gibbs RA, Daly MJ, Buxbaum JD, State MW, Devlin B, Roeder K (2013) Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet 9:e1003671
Hormozdiari F, Penn O, Borenstein E, Eichler EE (2015) The discovery of integrated gene networks for autism and related disorders. Genome Res 25:142–154
Huguet G, Bourgeron T (2016) The genetic causes of autism spectrum disorders. In: Synaptic dysfunction in autism spectrum disorder and intellectual disability (in press)
Huguet G, Ey E, Bourgeron T (2013) The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet 14:191–213
Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M (2008) Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci 28:1697–1708
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, Wigler M (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74:285–299
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221
Jacquemont ML, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois MC, Prieur M, Vekemans M, Carter NP, Munnich A, Colleaux L, Philippe A (2006) Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet 43:843–849
Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34:27–29
Jang SW, Liu X, Pradoldej S, Tosini G, Chang Q, Iuvone PM, Ye K (2010) N-acetylserotonin activates TrkB receptor in a circadian rhythm. Proc Natl Acad Sci USA 107:3876–3881
Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, Wang G, Liang J, Wang Z, Cao D, Carter MT, Chrysler C, Drmic IE, Howe JL, Lau L, Marshall CR, Merico D, Nalpathamkalam T, Thiruvahindrapuram B, Thompson A, Uddin M, Walker S, Luo J, Anagnostou E, Zwaigenbaum L, Ring RH, Wang J, Lajonchere C, Wang J, Shih A, Szatmari P, Yang H, Dawson G, Li Y, Scherer SW (2013) Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet 93:249–263
Kanner L (1943) Autistic disturbances of affective contact. Nerv Child 2:217–250
Kelleher RJ 3rd, Bear MF (2008) The autistic neuron: troubled translation? Cell 135:401–406
Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, Grice DE, Ledbetter DH, Lord C, Mane SM, Martin CL, Martin DM, Morrow EM, Walsh CA, Melhem NM, Chaste P, Sutcliffe JS, State MW, Cook EH Jr, Roeder K, Devlin B (2012) Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 3:9
Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K (2012) Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488:471–475
Krumm N, O’Roak BJ, Shendure J, Eichler EE (2014) A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 37:95–105
Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE (2015) Excess of rare, inherited truncating mutations in autism. Nat Genet 47:582–588
Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH Jr, Dobyns WB, Christian SL (2008) Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17:628–638
Lam KS, Aman MG, Arnold LE (2006) Neurochemical correlates of autistic disorder: a review of the literature. Res Dev Disabil 27:254–289
Le Couteur A, Bailey A, Goode S, Pickles A, Gottesman I, Robertson S, Rutter M (1996) A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry 37:785–801
Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, Konyukh M, Chaste P, Ey E, Rastam M, Anckarsäter H, Nygren G, Gillberg IC, Melke J, Toro R, Regnault B, Fauchereau F, Mercati O, Lemière N, Skuse D, Poot M, Holt R, Monaco AP, Järvelä I, Kantojärvi K, Vanhala R, Curran S, Collier DA, Bolton P, Chiocchetti A, Klauck SM, Poustka F, Freitag CM, Waltes R, Kopp M, Duketis E, Bacchelli E, Minopoli F, Ruta L, Battaglia A, Mazzone L, Maestrini E, Sequeira AF, Oliveira B, Vicente A, Oliveira G, Pinto D, Scherer SW, Zelenika D, Delepine M, Lathrop M, Bonneau D, Guinchat V, Devillard F, Assouline B, Mouren MC, Leboyer M, Gillberg C, Boeckers TM, Bourgeron T (2012) Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet 8:e1002521
Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, Giuliano F, Stordeur C, Depienne C, Mouzat K, Pinto D, Howe J, Lemière N, Durand CM, Guibert J, Ey E, Toro R, Peyre H, Mathieu A, Amsellem F, Rastam M, Gillberg IC, Rappold GA, Holt R, Monaco AP, Maestrini E, Galan P, Heron D, Jacquette A, Afenjar A, Rastetter A, Brice A, Devillard F, Assouline B, Laffargue F, Lespinasse J, Chiesa J, Rivier F, Bonneau D, Regnault B, Zelenika D, Delepine M, Lathrop M, Sanlaville D, Schluth-Bolard C, Edery P, Perrin L, Tabet AC, Schmeisser MJ, Boeckers TM, Coleman M, Sato D, Szatmari P, Scherer SW, Rouleau GA, Betancur C, Leboyer M, Gillberg C, Delorme R, Bourgeron T (2014) Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet 10:e1004580
Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, Buja A, Krieger A, Yoon S, Troge J, Rodgers L, Iossifov I, Wigler M (2011) Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70:886–897
Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H (2010) The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry 167:1357–1363
Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, Lek M, Liu L, Flannick J, Ripke S, Nagaswamy U, Muzny D, Reid JG, Hawes A, Newsham I, Wu Y, Lewis L, Dinh H, Gross S, Wang LS, Lin CF, Valladares O, Gabriel SB, dePristo M, Altshuler DM, Purcell SM, NHLBI Exome Sequencing Project, State MW, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Daly MJ (2013) Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77:235–242
Liu L, Sabo A, Neale BM, Nagaswamy U, Stevens C, Lim E, Bodea CA, Muzny D, Reid JG, Banks E, Coon H, Depristo M, Dinh H, Fennel T, Flannick J, Gabriel S, Garimella K, Gross S, Hawes A, Lewis L, Makarov V, Maguire J, Newsham I, Poplin R, Ripke S, Shakir K, Samocha KE, Wu Y, Boerwinkle E, Buxbaum JD, Cook EH Jr, Devlin B, Schellenberg GD, Sutcliffe JS, Daly MJ, Gibbs RA, Roeder K (2013) Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet 9:e1003443
Liu L, Lei J, Sanders SJ, Willsey AJ, Kou Y, Cicek AE, Klei L, Lu C, He X, Li M, Muhle RA, Ma'ayan A, Noonan JP, Sestan N, McFadden KA, State MW, Buxbaum JD, Devlin B, Roeder K (2014) DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism 5:22
Lundstrom S, Chang Z, Kerekes N, Gumpert CH, Rastam M, Gillberg C, Lichtenstein P, Anckarsater H (2011) Autistic-like traits and their association with mental health problems in two nationwide twin cohorts of children and adults. Psychol Med 41:2423–2433
Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10:307–318
Mabb AM, Ehlers MD (2010) Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol 26:179–210
Malow B, Adkins KW, McGrew SG, Wang L, Goldman SE, Fawkes D, Burnette C (2012) Melatonin for sleep in children with autism: a controlled trial examining dose, tolerability, and outcomes. J Autism Dev Disord 42:1729–1737, author reply 1738
Maronde E, Saade A, Ackermann K, Goubran-Botros H, Pagan C, Bux R, Bourgeron T, Dehghani F, Stehle JH (2011) Dynamics in enzymatic protein complexes offer a novel principle for the regulation of melatonin synthesis in the human pineal gland. J Pineal Res 51:145–155
Melke J et al (2008) Abnormal melatonin synthesis in autism spectrum disorders. Mol Psychiatry 13:90–98
Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, Wu W, Corominas R, Peoples A, Koren A, Gore A, Kang S, Lin GN, Estabillo J, Gadomski T, Singh B, Zhang K, Akshoomoff N, Corsello C, McCarroll S, Iakoucheva LM, Li Y, Wang J, Sebat J (2012) Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151:1431–1442
Miles JH, Takahashi TN, Bagby S, Sahota PK, Vaslow DF, Wang CH, Hillman RE, Farmer JE (2005) Essential versus complex autism: definition of fundamental prognostic subtypes. Am J Med Genet A 135:171–180
Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH (2013) Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol 12:406–414
Neale BM et al (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245
Nemirovsky SI, Cordoba M, Zaiat JJ, Completa SP, Vega PA, Gonzalez-Moron D, Medina NM, Fabbro M, Romero S, Brun B, Revale S, Ogara MF, Pecci A, Marti M, Vazquez M, Turjanski A, Kauffman MA (2015) Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS One 10:e0116358
O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE (2011) Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 43:585–589
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE (2012a) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250
O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O’Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J (2012b) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338:1619–1622
Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F (2001) Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell 105:257–267
Pagan C, Delorme R, Launay J, Bourgeron T (2012) Alterations of the serotonin-melatonin pathway in autism spectrum disorders: Biological evidence and clinical consequences. Oxford University Press, New York
Pagan C, Delorme R, Callebert J, Goubran-Botros H, Amsellem F, Drouot X, Boudebesse C, Le Dudal K, Ngo-Nguyen N, Laouamri H, Gillberg C, Leboyer M, Bourgeron T, Launay JM (2014) The serotonin-N-acetylserotonin-melatonin pathway as a biomarker for autism spectrum disorders. Transl Psychiatry 4:e479
Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G (2011) Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472:437–442
Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466:368–372
Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald JR, Mills R, Prasad A, Noonan K, Gribble S, Prigmore E, Donahoe PK, Smith RS, Park JH, Hurles ME, Carter NP, Lee C, Scherer SW, Feuk L (2011) Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 29:512–520
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, Vorstman JA, Thompson A, Regan R, Pilorge M, Pellecchia G, Pagnamenta AT, Oliveira B, Marshall CR, Magalhaes TR, Lowe JK, Howe JL, Griswold AJ, Gilbert J, Duketis E, Dombroski BA, De Jonge MV, Cuccaro M, Crawford EL, Correia CT, Conroy J, Conceição IC, Chiocchetti AG, Casey JP, Cai G, Cabrol C, Bolshakova N, Bacchelli E, Anney R, Gallinger S, Cotterchio M, Casey G, Zwaigenbaum L, Wittemeyer K, Wing K, Wallace S, van Engeland H, Tryfon A, Thomson S, Soorya L, Rogé B, Roberts W, Poustka F, Mouga S, Minshew N, McInnes LA, McGrew SG, Lord C, Leboyer M, Le Couteur AS, Kolevzon A, Jiménez González P, Jacob S, Holt R, Guter S, Green J, Green A, Gillberg C, Fernandez BA, Duque F, Delorme R, Dawson G, Chaste P, Café C, Brennan S, Bourgeron T, Bolton PF, Bölte S, Bernier R, Baird G, Bailey AJ, Anagnostou E, Almeida J, Wijsman EM, Vieland VJ, Vicente AM, Schellenberg GD, Pericak-Vance M, Paterson AD, Parr JR, Oliveira G, Nurnberger JI, Monaco AP, Maestrini E, Klauck SM, Hakonarson H, Haines JL, Geschwind DH, Freitag CM, Folstein SE, Ennis S, Coon H, Battaglia A, Szatmari P, Sutcliffe JS, Hallmayer J, Gill M, Cook EH, Buxbaum JD, Devlin B, Gallagher L, Betancur C, Scherer SW (2014) Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94:677–694
Risch N, Hoffmann TJ, Anderson M, Croen LA, Grether JK, Windham GC (2014) Familial recurrence of autism spectrum disorder: evaluating genetic and environmental contributions. Am J Psychiatry 171:1206–1213
Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM (1985) Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry 142:74–77
Ronald A, Hoekstra RA (2011) Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet 156B:255–274
Ronald A, Happe F, Plomin R (2005) The genetic relationship between individual differences in social and nonsocial behaviours characteristic of autism. Dev Sci 8:444–458
Ronald A, Happe F, Price TS, Baron-Cohen S, Plomin R (2006) Phenotypic and genetic overlap between autistic traits at the extremes of the general population. J Am Acad Child Adolesc Psychiatry 45:1206–1214
Ronald A, Larsson H, Anckarsater H, Lichtenstein P (2010) A twin study of autism symptoms in Sweden. Mol Psychiatry 16:1039–1047
Ronemus M, Iossifov I, Levy D, Wigler M (2014) The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet 15:133–141
Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA (2009) Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med 163:907–914
Rossignol DA, Frye RE (2012) A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry 17:389–401
Rutherford SL (2000) From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays 22:1095–1105
Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH Jr, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ (2014) A framework for the interpretation of de novo mutation in human disease. Nat Genet 46:944–950
Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, Mason CE, Bilguvar K, Celestino-Soper PB, Choi M, Crawford EL, Davis L, Wright NR, Dhodapkar RM, DiCola M, DiLullo NM, Fernandez TV, Fielding-Singh V, Fishman DO, Frahm S, Garagaloyan R, Goh GS, Kammela S, Klei L, Lowe JK, Lund SC, McGrew AD, Meyer KA, Moffat WJ, Murdoch JD, O’Roak BJ, Ober GT, Pottenger RS, Raubeson MJ, Song Y, Wang Q, Yaspan BL, Yu TW, Yurkiewicz IR, Beaudet AL, Cantor RM, Curland M, Grice DE, Günel M, Lifton RP, Mane SM, Martin DM, Shaw CA, Sheldon M, Tischfield JA, Walsh CA, Morrow EM, Ledbetter DH, Fombonne E, Lord C, Martin CL, Brooks AI, Sutcliffe JS, Cook EH Jr, Geschwind D, Roedr K, Devlin B, State MW (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70:863–885
Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Günel M, Roeder K, Geschwind DH, Devlin B, State MW (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485:237–241
Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A (2014) The familial risk of autism. JAMA 311:1770–1777
Sando R 3rd, Gounko N, Pieraut S, Liao L, Yates J 3rd, Maximov A (2012) HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151:821–834
Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E (2013) Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 493:411–415
Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen AL, Udvardi PT, Shiban E, Spilker C, Balschun D, Skryabin BV, Dieck S, Smalla KH, Montag D, Leblond CS, Faure P, Torquet N, Le Sourd AM, Toro R, Grabrucker AM, Shoichet SA, Schmitz D, Kreutz MR, Bourgeron T, Gundelfinger ED, Boeckers TM (2012) Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 486:256–260
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449
Sheng M, Kim E (2011) The postsynaptic organization of synapses. Cold Spring Harb Perspect Biol 3
Shi L, Zhang X, Golhar R, Otieno FG, He M, Hou C, Kim C, Keating B, Lyon GJ, Wang K, Hakonarson H (2013) Whole-genome sequencing in an autism multiplex family. Mol Autism 4:8
Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN (2011) Sociability and motor functions in Shank1 mutant mice. Brain Res 1380:120–137
Skuse DH, Mandy W, Steer C, Miller LL, Goodman R, Lawrence K, Emond A, Golding J (2009) Social communication competence and functional adaptation in a general population of children: preliminary evidence for sex-by-verbal IQ differential risk. J Am Acad Child Adolesc Psychiatry 48:128–137
Sompol P, Liu X, Baba K, Paul KN, Tosini G, Iuvone PM, Ye K (2011) N-acetylserotonin promotes hippocampal neuroprogenitor cell proliferation in sleep-deprived mice. Proc Natl Acad Sci USA 108:8844–8849
Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiat All Discip 30:405–416
Szafranski P, Schaaf CP, Person RE, Gibson IB, Xia Z, Mahadevan S, Wiszniewska J, Bacino CA, Lalani S, Potocki L, Kang SH, Patel A, Cheung SW, Probst FJ, Graham BH, Shinawi M, Beaudet AL, Stankiewicz P (2010) Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum Mutat 31:840–850
Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Rogé B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bölte S, Feineis-Matthews S, Herbrecht E, Schmötzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ (2007) Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39:319–328
Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S (2008) Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet 147B:844–849
Tordjman S, Anderson GM, Pichard N, Charbuy H, Touitou Y (2005) Nocturnal excretion of 6-sulphatoxymelatonin in children and adolescents with autistic disorder. Biol Psychiatry 57:134–138
Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, Coleman M, Leboyer M, Gillberg C, Bourgeron T (2010) Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet 26:363–372
Toro R, Poline JB, Huguet G, Loth E, Frouin V, Banaschewski T, Barker GJ, Bokde A, Büchel C, CarvalhoFM CP, Fauth-Bühler M, Flor H, Gallinat J, Garavan H, Gowland P, Heinz A, Ittermann B, Lawrence C, Lemaître H, Mann K, Nees F, Paus T, Pausova Z, Rietsche M, Robbins T, Smolka MN, Ströhle A, Schumann G, Bourgeron T (2014) Genomic architecture of human neuroanatomical diversity. Mol Psychiatry 20:1011–1016
Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, Pinto D, Lau L, Nalpathamkalam T, Marshall CR, Blencowe BJ, Frey BJ, Merico D, Yuen RK, Scherer SW (2014) Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet 46:742–747
Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474:380–384
Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, Franke L (2006) Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry 11(1):18–28
Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH (2011) Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet 20:3093–3108
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ, Autism Consortium (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358:667–675
Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, Murtha MT, Bichsel C, Niu W, Cotney J, Ercan-Sencicek AG, Gockley J, Gupta AR, Han W, He X, Hoffman EJ, Klei L, Lei J, Liu W, Liu L, Lu C, Xu X, Zhu Y, Mane SM, Lein ES, Wei L, Noonan JP, Roeder K, Devlin B, Sestan N, State MW (2013) Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155:997–1007
Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN (2011) Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One 6:e20631
Won H, Lee H-R, Gee HY, Mah W, Kim J-I, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park S-G, Lee J-S, Lee K, Kim D, Bae YC, Kaang B-K, Lee MG, Kim E (2012) Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486:261–265
Wright B, Sims D, Smart S, Alwazeer A, Alderson-Day B, Allgar V, Whitton C, Tomlinson H, Bennett S, Jardine J, McCaffrey N, Leyland C, Jakeman C, Miles J (2011) Melatonin versus placebo in children with autism spectrum conditions and severe sleep problems not amenable to behaviour management strategies: a randomised controlled crossover trial. J Autism Dev Disord 41:175–184
Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, Tyler-Smith C, Genomes Project C (2012) Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 91:1022–1032
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42:565–569
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Yang M, Bozdagi O, Scattoni ML, Woehr M, Roullet FI, Katz AM, Abrams DN, Kalikhman D, Simon H, Woldeyohannes L, Zhang JY, Harris MJ, Saxena R, Silverman JL, Buxbaum JD, Crawley JN (2012) Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci 32:6525–6541
Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, D’Gama AM, Lim ET, Sanders SJ, Mochida GH, Partlow JN, Sunu CM, Felie JM, Rodriguez J, Nasir RH, Ware J, Joseph RM, Hill RS, Kwan BY, Al-Saffar M, Mukaddes NM, Hashmi A, Balkhy S, Gascon GG, Hisama FM, LeClair E, Poduri A, Oner O, Al-Saad S, Al-Awadi SA, Bastaki L, Ben-Omran T, Teebi AS, Al-Gazali L, Eapen V, Stevens CR, Rappaport L, Gabriel SB, Markianos K, State MW, Greenberg ME, Taniguchi H, Braverman NE, Morrow EM, Walsh CA (2013) Using whole-exome sequencing to identify inherited causes of autism. Neuron 77:259–273
Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y, Gazzellone MJ, D’Abate L, Deneault E, Howe JL, Liu RS, Thompson A, Zarrei M, Uddin M, Marshall CR, Ring RH, Zwaigenbaum L, Ray PN, Weksberg R, Carter MT, Fernandez BA, Roberts W, Szatmari P, Scherer SW (2015) Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 21:185–191
Acknowledgments
We thank Roberto Toro for critical reading of the manuscript. This work was funded by the Institut Pasteur, the Bettencourt-Schueller foundation, Centre National de la Recherche Scientifique, University Paris Diderot, Agence Nationale de la Recherche (SynDiv-ASD), the Conny-Maeva Charitable Foundation, the Cognacq Jay Foundation, the Orange Foundation, and the Fondamental Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-Noncommercial 2.5 License (http://creativecommons.org/licenses/by-nc/2.5/) which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2016 The Author(s)
About this chapter
Cite this chapter
Huguet, G., Benabou, M., Bourgeron, T. (2016). The Genetics of Autism Spectrum Disorders. In: Sassone-Corsi, P., Christen, Y. (eds) A Time for Metabolism and Hormones. Research and Perspectives in Endocrine Interactions. Springer, Cham. https://doi.org/10.1007/978-3-319-27069-2_11
Download citation
DOI: https://doi.org/10.1007/978-3-319-27069-2_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27068-5
Online ISBN: 978-3-319-27069-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)