Abstract
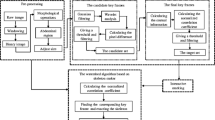
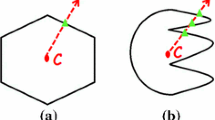
X-ray images segmentation can be useful to aid in accurate diagnosis or faithful 3D bone reconstruction but remains a challenging and complex task, particularly when dealing with large and complex anatomical structures such as the human pelvic bone. In this paper, we propose a multi-atlas fusion framework to automatically segment the human pelvic structure from 45 or 135-degree oblique X-ray radiographic images. Unlike most atlas-based approach, this method combines a data set of a priori segmented X-ray images of the human pelvis (or multi-atlas) to generate an adaptive superpixel map in order to take efficiently into account both the imaging pose variability along with the inter-patient (bone) shape non-linear variability. In addition, we propose a new label propagation or fusion step based on the variation of information criterion for integrating the multi-atlas information into the final consensus segmentation. We thoroughly evaluated the method on 30 manually segmented 45 or 135 degree oblique X-ray radiographic images data set by performing a leave-one-out study. Compared to the manual gold standard segmentations, the accuracy of our automatic segmentation approach is \(85\%\) which remains in the error range of manual segmentations due to the inter intra/observer variability.
Chapter PDF
Similar content being viewed by others
Keywords
References
Mignotte, M., Collet, C., Pérez, P., Bouthemy, P.: Sonar image segmentation using an unsupervised hierarchical MRF model. IEEE Trans. on Image Processing 9(7), 1216–1231 (2000)
Mignotte, M., Meunier, J., Tardif, J.-C.: Endocardial boundary estimation and tracking in echocardiographic images using deformable templates and markov random fields. Pattern Analysis and Applications 4(4), 256–271 (2001)
Mignotte, M., Meunier, J.: A multiscale optimization approach for the dynamic contour-based boundary detection issue. Computerized Medical Imaging and Graphics 25(3), 265–275 (2001)
Destrempes, F., Mignotte, M.: Localization of shapes using statistical models and stochastic optimization. IEEE Trans. on Pattern Analysis and Machine Intelligence 29(9), 1603–1615 (2007)
Artaechevarria, X., Muñoz-Barrutia, A., Ortiz-de-Solorzano, C.: Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Trans. Med. Imaging 28(8), 1266–1277 (2009)
Aljabar, P., Heckemann, R.A., Hammers, A., Hajnal, J.V., Rueckert, D.: Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage, 726–738 (2009)
Dowling, J.A., Fripp, J., Chandra, S., Pluim, J.P.W., Lambert, J., Parker, J., Denham, J., Greer, P.B., Salvado, O.: Fast automatic multi-atlas segmentation of the prostate from 3D MR images. In: Madabhushi, A., Dowling, J., Huisman, H., Barratt, D. (eds.) Prostate Cancer Imaging 2011. LNCS, vol. 6963, pp. 10–21. Springer, Heidelberg (2011)
Morin, J.-P., Desrosiers, C., Duong, L.: A random walk approach for multiatlas-based segmentation. In: ICPR 2012, pp. 3636–3639 (2012)
Rohlfing, T., Brandt, R., Menzel, R., Maurer, C.R.: Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage 21(4), 1428–1442 (2004)
Rohlfing, T., Russakoff, D.B., Maurer, C.R.: Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans. Med. Imaging 23(8), 983–994 (2004)
Mignotte, M.: A label field fusion model with a variation of information estimator for image segmentation. Information Fusion 20, 7–20 (2014)
Mignotte, M., Meunier, J., Soucy, J.-P.: DCT-based complexity regularization for EM tomographic reconstruction. IEEE Trans. on Biomedical Engineering 55(2), 801–805 (2008)
Yu, G., Sapiro, G.: DCT image denoising: a simple and effective image denoising algorithm. Image Processing On Line 1 (2011)
Canny, J.: A computational approach to edge detection. IEEE Transactions on Pattern Analysis and Machine Intelligence 8(6), 679–698 (1986)
Benameur, S., Mignotte, M., Parent, S., Labelle, H., Skalli, W., De Guise, J.: 3d/2d registration and segmentation of scoliotic vertebrae using statistical models. Computerized Medical Imaging and Graphics 27(5), 321–327 (2003)
Ren, X., Malik, J.: Learning a classification model for segmentation. In: 9th IEEE International Conference on Computer Vision, vol. 1, pp. 10–17 (October 2003)
Jodoin, P.-M., Mignotte, M., Rosenberger, C.: Segmentation framework based on label field fusion. IEEE Trans. on Image Processing 16(10), 2535–2550 (2007)
Mignotte, M.: A segmentation-based regularization term for image deconvolution. IEEE Trans. on Image Processing 15(7), 1973–1984 (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Nguyen, D.C.T., Benameur, S., Mignotte, M., Lavoie, F. (2015). Superpixel and Entropy-Based Multi-atlas Fusion Framework for the Segmentation of X-ray Images. In: Murino, V., Puppo, E. (eds) Image Analysis and Processing — ICIAP 2015. ICIAP 2015. Lecture Notes in Computer Science(), vol 9280. Springer, Cham. https://doi.org/10.1007/978-3-319-23234-8_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-23234-8_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23233-1
Online ISBN: 978-3-319-23234-8
eBook Packages: Computer ScienceComputer Science (R0)