Abstract
This chapter provides a general explanation of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). The SHIME is one of the few gut models that mimics the entire gastrointestinal tract incorporating stomach, small intestine and different colon regions. After a general description of the model’s development history and an overview of the specific features that distinguish SHIME from other gut models, we will give some insight in the general protocol of running SHIME experiments. However, with the SHIME being a highly flexible experimental setup, we also dedicate some part of this chapter discussing the modifications that can be performed, especially with respect to simulation of the mucosal (micro-)environment and interaction with host cells.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Description of SHIME®
‘SHIME’ is an acronym for the Simulator of the Human Intestinal Microbial Ecosystem and since 2010, the name has been jointly registered by ProDigest and Ghent University. This paragraph primarily describes the conventional experimental setup of the SHIME® system. Yet, because of its modular setup, the SHIME is highly flexible and it can be technically modified to target digestive conditions of interest: this will be briefly discussed further down this chapter.
1.1 History of the Model
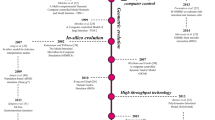
The Simulator of the Human Intestinal Microbial Ecosystem (SHIME) is a multi-compartment dynamic simulator of the human gut developed in 1993 (Molly et al. 1993). The development of multi-compartment simulators of (parts of) the human gut originated from the awareness that fecal microbiota significantly differ from the in vivo colon microbiota in terms of community composition and metabolic activity. While inoculation of fecal microbiota into single-stage chemostats was a first attempt to mimic colon conditions, it was only useful for limited periods of time since environmental parameters such as pH, redox potential, available nutrients and microbial population dynamics constantly change. In order to maintain the inoculated intestinal microbiota over a longer timeframe, semi-continuous fermenters were developed where the intermittent supplementation of nutritional medium and the removal of microbial suspension could be simulated (Miller and Wolin 1981). While the latter systems typically make use of one single fermenter, the colon is a very heterogeneous region with clear differences in substrate availability, fermentation activity, microbial composition and several environmental conditions. This makes it impossible to simulate a representative culture of colon microbiota in one compartment. Several multi-compartment reactors were therefore developed to simulate the different conditions of the colon lumen (Macfarlane et al. 1989; Miller and Wolin 1981), from which the SHIME was one of the last in this generation of gut simulators.
Technically, the SHIME is an evolution of the simulator of the University of Reading introduced by Macfarlane et al. (1989) and mimics the conditions in the ascending, transverse and descending colon regions. The SHIME differentiates from the Reading model by incorporation of upper digestive tract conditions, leading to a succession of five compartments simulating the upper (stomach, small intestine) and the lower (ascending, transverse and descending colon) digestive tract.
The entire SHIME reactor operates at 37 °C. It contains double-jacketed glass vessels that are connected through peristaltic pumps (Fig. 27.1). The first two reactors follow a fill-and-draw principle adding three times a day a defined nutritional medium to a gastric compartment, and pancreatic and bile liquid to a small intestine compartment. The medium is composed of complex carbohydrate and protein sources with addition of mucins and a mineral and vitamin mix (Molly et al. 1993). Upon digestion in the gastric and intestine compartments, the slurry is pumped in the ascending colon vessel where colon digestion is initiated. The three colon compartments are continuously stirred with constant volume and pH control. Retention times in the upper digestive tract can be modulated by changing the flow rates from the gastric and intestine compartments, while retention times from the colon compartments are primarily modulated through a change in compartment volume. Depending on the human target group of interest the retention time may vary from 24 h to 72 h.
The pH of the gastric compartment used to work at a fixed pH of 2.0, yet with the advent of a completely computer-controlled SHIME system, specific pH profiles during gastric and intestine digestion can be established as well. While the small intestine compartment typically operates at slightly acidic to neutral conditions, the pH of the colon compartments is controlled between 5.6 and 5.9 in the ascending, 6.1–6.4 in the transverse and 6.6–6.9 in the descending colon. Mixing of the digestive slurry in the respective compartments is obtained with magnetic stir bars. The entire SHIME system is kept anaerobic by daily flushing the headspace of the respective compartments with N2 gas or a 90/10 % N2/CO2 gas mixture.
1.2 Special Features of the Model
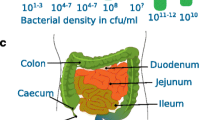
The emphasis of the SHIME® system is primarily put on the simulation of the colon microbial community. Because of the inaccessibility of the human colon region to take a representative microbial inoculum, the fecal microbiota is chosen as inoculum to the colon compartments of the SHIME reactor. The fecal microbiome is significantly different from the in vivo colon microbiome, both in terms of composition as metabolic activity. Yet, the colon being considered as a plug-flow system, the fecal microbiome is nothing else than a colon microbiome that has undergone community and metabolic shifts during transit from the proximal colon to the rectum. The idea of the SHIME system—and other multi-stage colon compartment reactors such as the Reading model—is to allow a suitable adaptation period for the fecal microbiome to adapt to the conditions that prevail in the respective colon compartments. From an engineering perspective a suitable adaptation time for starting up a reactor is around 5–10 times its residence time. Taking the example of a male individual with a gut residence time of 48 h and imposing that in the SHIME system, this would entail an adaptation time of 10–20 days upon inoculation with the fecal microbiota in the colon compartments. We will come back to this stabilization aspect in the next paragraph.
A second aspect is the choice of inoculum. The SHIME is typically inoculated with the fecal microbiome, derived from one individual. There has been or there still is a lot of debate on what is the most suitable inoculum for mimicking the human gut microbiome in the most representative way. Some research groups specifically opt for pooling fecal microbiota from for example ten different individuals (Minekus et al. 1999), thereby (partly) accounting for the huge interindividual variability that exists in microbiome composition and to incorporate properties from different microbiomes in order to create an ‘average’ 11th microbiome. Given the enormous functional redundancy of the gut microbiome such pooled microbiome will indeed take on a normal fermentation profile which generally not that different from the microbial fermentation profile of a single individual. While such approach may work when investigating hydrolysis and fermentation of carbohydrates, it fails to accurately mimic microbial processes that lack this functional redundancy. To exemplify, it has become clear that the microbial metabolic potency towards polyphenols such as daidzein, isoxanthohumol, catechins and others is highly dependent on an individual’s microbiome (van Duynhoven et al. 2011). The existence of different ‘metabotypes’ has therefore even been proposed to distinguish a bioactive metabolite producing phenotype from a non-producing phenotype (Bolca et al. 2013). The above element is the primary reason why a SHIME reactor is inoculated with the fecal microbiome from one individual and succeeds in maintaining the microbial metabolic phenotype towards specific polyphenols during the in vivo/in vitro transition and several weeks after (Decroos et al. 2006; Possemiers et al. 2006).
The choice for individual inocula also appears to be a crucial aspect in recent studies where the SHIME was fundamentally optimized to enable colonization of the mucosal microbiome (Van den Abbeele et al. 2013). This concerns a third special feature of the SHIME model. The mucosal microbiome is this part of the gut microbial ecosystem that is able to colonize the mucus overlying the gut epithelium. Due to its close proximity to host epithelial cells, the mucosal microbiome is thought to have an intrinsically higher potency to modulate gut health, and by extension, human health (Van den Abbeele et al. 2011). The mucosal microbiome was already known to fundamentally differ from the luminal microbiome in composition and interestingly, presence of important mucosal colonizers such as Faecalibacterium prausnitzii seems to negatively correlate with occurrence (Willing et al. 2009) and postoperative recurrence of ileal Crohn’s disease (Sokol et al. 2008). Given the difficult access to the mucosal environment, the development of gut simulators that accurately mimic mucosal microbial colonization is considered a strong asset to obtain a better understanding of the host-microbe interactome. It is in this philosophy that Van den Abbeele et al. (2013) decided to optimize the SHIME for mimicking mucosal microbial colonization by incorporation of mucin-covered microcosms. The major finding of this so-called M-SHIME, or mucosal SHIME, was that colonization of the mucosal environment was characterized by a higher abundance of butyrate producing Clostridium clusters IV and XIVa. This phylogenetic group is considered crucial for delivering butyrate as primary energy source to colonocytes and improves gut barrier function by strengthening the tight junctions. Coming back to the importance of individual inocula and avoiding pooled samples, it was also demonstrated that the M-SHIME was able to maintain the unique features of an individual’s microbiome in terms of its mucosal composition (Van den Abbeele et al. 2013).
The fourth feature is the flexibility of the SHIME model and the ease with which reactor compartments can be added or left away. This modular setup is useful when a placebo-controlled study needs to be conducted, when different prebiotics need to be compared (Grootaert et al. 2009) or when a microbiome phenotype producing a bioactive metabolite is compared with a non-producing phenotype (Possemiers et al. 2006). Moreover, it is even possible to explore the interindividual variability in microbiome behavior upon specific treatments by having a common upper digestive tract simulation in the gastric and intestine compartments and subsequently split up the system in several parallel colon compartments, each of which are inoculated with the fecal microbiota from separate individuals.
A fifth feature of the SHIME model refers to the possibility of simulating the microbiome from different human target groups such as adult vs. infant, healthy vs. diseased (e.g. ulcerative colitis patients: Vermeiren et al. 2012) as well as the simulation of animal (pig, dog) microbiomes. In each of these specific cases, the microbial inocula, residence times of the different gastric compartments, composition of the gastric juices, region specific pH’s, feed, feeding regimes and body temperatures are adapted in the SHIME set-up leading to an accurate and relevant simulation of the targeted human or animal host. Finally, other features of the SHIME include the gradual emptying of the gastric digest into the intestine compartment, the option of running a dynamic pH profiles in the gastric compartment and the possibility of putting a dialysis unit behind the intestine compartment to enable running experiments with real food matrices or food constituents that need to undergo predigestion and removal of sugar monomers or amino acids and peptides before the digest is transferred to the colon compartment.
1.3 Stability and Reproducibility of the System
As the SHIME® reactor is inoculated with a fecal microbiome, the latter needs an appropriate amount of time to adapt to the prevailing environmental conditions in the respective colon compartments. This adaptation process was studied more in detail by Possemiers et al. (2004) who monitored the increasing colonization of microbial groups of interest upon inoculation, as well as their metabolic activity. Highly-abundant groups such as Bacteroides obtained stable concentrations more easily, at 10 days, compared to less-abundant groups such as lactobacilli which needed 15 days of stabilization. To obtain a stable functionality in terms of short chain fatty acid production, an adaptation period of at least 15 days, even approaching 20 days, was needed. The length of stabilization obviously relates to the residence time that is imposed in the SHIME, but may also depend on the microbiome composition as such.
As the SHIME is a highly standardized system, and many digestive parameters are under control of the operator, it also leads to highly reproducible results. This is especially required when different products such as novel prebiotics, candidate drug components or new plant extracts, need to be compared with one another. When it is not always possible to run different SHIMEs in parallel, for example because of the multitude of test compounds, SHIME experiments may need to be repeated. The scientist in charge needs to be cautious to conduct this experiment in exactly the same way and with the same inoculum to enable an adequate comparison between different compounds. Such reproducibility was previously tested for the conventional SHIME (without mucin-coated microcosms) and proven very effective in obtaining a reproducible microbial colonization process and accompanied metabolic activity (Van den Abbeele et al. 2010). Noteworthy, the authors found a small preferential colonization of Bacteroidetes and Clostridium cluster IX in the colon compartments in comparison with the stool sample. While the overall colonization profile was still representative of the in vivo colonization process, this slight bias has been a common observation for several dynamic models that work with luminal content only. Van den Abbeele et al. (2013) succeeded in removing this colonization selectivity by incorporating mucin-covered microcosms in the SHIME, thereby creating the M-SHIME or mucosal SHIME. Butyrate producing Clostridium clusters IV and XIVa were found to specifically colonize the mucosal environment thereby compensating for their lower abundance in the lumen.
1.4 Relevance to Human In Vivo Situation
The first validation paper of the conventional SHIME® setup concerned a study from Molly et al. (1994) where several microbe-associated characteristics were defined. The authors focused on fermentation profiles of pectin, xylan, arabinogalactan and starch and found these to be consistent with the results from incubations of the same products with fecal microbiota from human volunteers (Englyst et al. 1987). Similarly, an enzymatic profile focusing on glycosyl hydrolases (galactosidase, glucosidase, xylosidase) was recorded and no differences with the fecal incubations were found. Apart from the comparison with fermentation activity the metabolic potency towards sulphasalazin, a prodrug for ulcerative colitis treatment, and its conversion to the active compound 5-aminosalicylic acid (5-ASA) was also evaluated. Consistent with in vivo literature, no 5-ASA was detected in the gastric compartment, while small amounts of 5-ASA were detected in the intestine compartment and full sulphasalazin conversion was observed in the colon compartments (Peppercorn and Goldman 1972).
A second validation study discusses the preservation of metabolic phenotypes when transferring fecal microbiota from an individual to the in vitro SHIME system. Focusing on the microbial conversion of isoxanthohumol to 8-prenylnaringenin (8-PN), a metabolite with strong pseudo-estrogenic activity, Possemiers et al. (2006) demonstrated a one-on-one correlation between urinary 8-PN excretion by human individuals and their microbiome. Secondly, the microbiome from a 8-PN producing and non-producing individual could be stably transferred to the SHIME system and the metabolic phenotype was adequately preserved.
A third validation study concerns the application of the M-SHIME where both the luminal as mucosal microbiome from an individual were simulated in the SHIME colon compartment (Van den Abbeele et al. 2013). Analyzing the microbiome from the luminal and mucosal regions with the HIT-Chip micro-array revealed the largest variability in the microbial dataset to originate from the in vivo/in vitro transition and difference between the luminal and mucosal environment. Yet, those species that account for the unique profile of an individual’s microbiome were preserved in the M-SHIME as well and accounted for 25 % of the variability in the microbial dataset.
1.5 Quality in Relation to Other Models with the Same Applicability
The SHIME® model is the sole in vitro model that integrates the entire gastrointestinal transit into one system. This is interesting to study for example digestibility of prebiotic substrates and its subsequent fermentability in the colon or the survival of pathogens or probiotics in the upper digestive tract before they reach the colon environment. Yet, one must consider that the conventional SHIME system operates without an absorption unit: this means that the nutritional medium for the SHIME must already be deprived of easily digestible carbohydrates or proteins that would normally be absorbed in the intestine.
The SHIME is the last of a generation of multi-compartment models that operate according to a semi-continuous stirred tank reactors setup. In comparison with the Reading model (Macfarlane et al. 1989) the SHIME connects the different compartments through peristaltic pumps. In terms of studying the gut microbiome over a long timeframe, both models would however be applicable.
In general, it must be stressed that the SHIME model has a strong emphasis on the ecological aspects of the colon microbiome. This entails that incubation experiments with the SHIME reactor are seldomly short and commonly take several weeks. This is put in place to look at the gradual adaptation of the microbiome to incoming substrates of interest (e.g. prebiotics or pharmaceuticals) or to evaluate the resilience of the microbial community against colonization by a (opportunistic) pathogen. Such research, which often necessitates an experimental period of more than a week to several weeks, strongly differs from the TIM-2 model, which monitors the microbiome on a shorter timeframe. One could conclude that short term experiments are highly suitable for evaluating the immediate metabolic potency of a carbohydrate or colonization ability of a microorganism of interest, while long-term experiments are primarily suitable to look at the adaptation of the microbial ecosystem to changing environmental conditions or inputs.
A last strong asset of the SHIME is its extension to M-SHIME—incorporating the mucosal microbiota (Van den Abbeele et al. 2013). While previous attempts primarily focused on the immediate and aspecific adhesion potency of gut microorganisms to mucin-covered glass slides, the integration of mucin-covered microcosms that can be replaced, to mimic desquamation, has been a breakthrough in the simulation of the mucosal microbiome and in the understanding of its dynamics.
2 General Protocol
Inoculation of the SHIME® colon compartments occurs with microbiota that has been isolated from fecal material of one individual. In contrast with other gut models, we deliberately choose not to work with the microbiome from pooled fecal samples from different human volunteers. The artificially high microbial diversity in pooled inocula creates disturbances in the cross-feeding processes between microorganisms that are adapted to one another in each of the separate microbiomes. We therefore advise to study interindividual variability through separate experiments. Upon inoculation of the colon compartments with fecal microbiota, the microbiome is given time to adapt itself to the prevailing conditions in the ascending, transverse and descending colon compartments.
A typical SHIME experiment consists of four stages: a stabilization period (2 weeks) to allow adaptation of the microbial community to the environmental conditions in the respective colon regions; a basal period (2 weeks) in which the reactor is operated under nominal conditions and baseline parameters are measured; a treatment period (2–4 weeks) where the effect of a specific treatment on the gastrointestinal microbial community is tested; and a washout period (2 weeks) to determine how long the changes induced by the tested substance can still be measured in the absence of the substance itself. This approach has mainly been used to investigate the activity and stability of probiotics and prebiotics during gastrointestinal transfer, the microbial conversion of bioactive food components (e.g. phytoestrogens), the metabolism of pharmaceutical components, the efficacy of colonic targeted delivery systems and the conversion and biological (in)activation of food and/or ingested environmental contaminants.
Shorter-term SHIME experiments are also a possibility. Specifically in the context of monitoring the initial stages of microbial colonization on the mucosal surface, 1-week experiments can be conducted. Distinguishing the mucosal from the luminal microbiome, Van den Abbeele et al. (2013) evaluated the colonization process of the microbiome derived from five human volunteers in M-SHIME systems over a timeframe of 5 days. The presence of mucins seems to play a specific role in the colonization process. In contrast with mucin surfaces, other contact surfaces, such as agar or plastic surfaces, did not result in a distinct colonization profile. Despite the fact that the applied mucins are derived from the porcine gut—and hence do not have the exact same composition as human mucins—the presence of this fairly similar glycoprotein surface already is an important driver in the colonization process.
Finally, by choosing the inoculum, tweaking digestive parameters, incorporating surface carriers or working with parallel reactor compartments, a SHIME operator is able to mimic the gastrointestinal conditions of a host target group or phenotype of interest, to incorporate internal control experiments or experimental replicates and to take into account the gastrointestinal colonization by mucosal microbiota.
3 Controls to Test Stability and Performance of the Model
As the SHIME® system can be operated over a longer timeframe, stability of the system is a crucial aspect. Investigating the modulation of the intestinal microbiome through certain treatments, also requires proper knowledge of a baseline situation and assumption of a stable microbiome. The microbial community composition and fermentation activity in the respective colon compartments is therefore closely monitored, especially during reactor startup, to evaluate the reactor’s capacity to create a stable microbiome that is still representative for the human in vivo situation. Using moving window correlation, Possemiers et al. (2004) previously introduced a stability criterion based on measurements with PCR-DGGE for different microbial groups, short chain fatty acids and ammonium. Calculating the correlation between two consecutive days along reactor startup, the authors considered a community to be stable once 80 % correlation was measured. The remaining 15–20 % variability typically originates from normal biological fluctuations. Overall, community stability is reached after about 2 weeks, while functional stability is obtained after 3 weeks. This startup phase seems quite long for a conventional SHIME experiment, but it is desired when the microbiome from different colon regions is under study and when the research question concerns microbial adaptations to applied treatments. Yet, short-term SHIME experiments can be conducted as well. The mucosal colonization process in the M-SHIME occurs quite rapidly—within 5 days—when a clear distinction between luminal and mucosal microbiome composition is noted. The preferential colonization of butyrate producing clostridia in the mucosal environment is one of the characteristic features (Van den Abbeele et al. 2013).
4 Read-Out of the System and How This Information Can Be Used
Assessing fermentation activity is one of the most important SHIME® read-outs. During operation of the SHIME system, short chain fatty acid or ammonia production can be preliminary assessed by the amount of NaOH or HCl that is supplemented by the pH controllers to maintain proper pH values in the respective colon vessels. As the conventional setup of the SHIME system does not have an absorption unit, short chain fatty acids (SCFA) do accumulate throughout the distal colon compartments. However, the SCFA concentrations in the SHIME do not lead to values above 100 mM, which would eventually result in product inhibition of the fermentation process. As saccharolytic conditions are more abundant than proteolytic conditions in the proximal colon and vice versa, the operator can still deduce the drop in saccharolytic activity by subtracting the SCFA concentration in the proximal colon from the SCFA concentration in the distal colon and thereby obtain the net amount of SCFA that are produced in the distal colon. Such strategy has been used before to discern the colon regions where specific fibers or oligosaccharides are broken down (Grootaert et al. 2009).
While adaptation of microbiome fermentation profiles to changing nutritional conditions is often monitored over several weeks (Grootaert et al. 2009), the immediate metabolic potency of the colon microbiome towards specific compounds of interest can be evaluated within days, e.g. with the onset of isoxanthohumol conversion into 8-prenylnaringenin (Possemiers et al. 2006), or even hours, e.g. with the microbial conversion of tea catechins and wine polyphenols (Gross et al. 2010).
Apart from the general fermentation activity and metabolic potency, the SHIME is evaluated for the microbiome composition in the respective colon vessels. The common approach typically includes establishment of DGGE profiles, either at Eubacteria or at group-specific level, as a quick screening for evaluating what treatments or what sample times are interesting to study in more detail. This can then be complemented by a quantitative analysis with q-PCR or a high-throughput analysis at the phylogenetic level with next generation sequencing. One of the important read-outs with microbiome fingerprints and profiles is the determination of the so-called ‘MRM-parameters’ (microbial resource management) (Marzorati et al. 2009). These typically contain calculation of richness, evenness and diversity indices and of microbiome dynamics throughout the study period. Especially evenness seems to be a strong indicator of microbiome resilience against invasion by exogenous species (Wittebolle et al. 2009).
A final read-out consists of an evaluation of host-microbe interactions. Colon suspension or the derived intestinal water (supernatant after centrifugation) can be brought in indirect or direct contact with host epithelial cells. This allows assessing to what extent changes in microbiome composition, microbial metabolites, signaling molecules or antigens have differential effects at the level of the host. This can be performed both with epithelial cells (Grootaert et al. 2011) as with a combination of epithelial and immune cells (Possemiers et al. 2013). Interestingly, the SHIME has recently been coupled to the Host-Microbe Interaction Module (HMI), which is a bi-compartmental system containing mucosal microbiota one the one side and host cells on the other side of a semi-permeable membrane (Marzorati et al. 2014).
5 Advantages, Disadvantages and Limitations of the System
The advantages, disadvantages and limitations of the system are summarized in Table 27.1.
References
Bolca S, Van de Wiele T, Possemiers S (2013) Gut metabotypes govern health effects of dietary polyphenols. Curr Opin Biotechnol 24:220–225
Ceuppens S, Uyttendaele M, Drieskens K, Heyndrickx M, Rajkovic A, Boon N et al (2012) Survival and germination of Bacillus cereus spores without outgrowth or enterotoxin production during in vitro simulation of gastrointestinal transit. Appl Environ Microbiol 78:7698–7705
Decroos K, Eeckhaut E, Possemiers S, Verstraete W (2006) Administration of equol-producing bacteria alters the equol production status in the simulator of the gastrointestinal microbial ecosystem (SHIME). J Nutr 136:946–952
Englyst HN, Hay S, MacFarlane GT (1987) Polysaccharide breakdown by mixed populations of human fecal bacteria. FEMS Microbiol Ecol 95:163–171
Grootaert C, Van den Abbeele P, Marzorati M, Broekaert WF, Courtin CM, Delcour JA et al (2009) Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 69:231–242
Grootaert C, Van de Wiele T, Van Roosbroeck I, Possemiers S, Vercoutter-Edouart A, Verstraete W, Bracke M, Vanhoecke B (2011) Bacterial monocultures, propionate, butyrate and H2O2 modulate the expression, secretion and structure of the fasting induced adipose factor in gut epithelial cells. Environ Microbiol 13:1778–1789
Gross G, Jacobs D, Peters S, Possemiers S, van Duynhoven J, Vaughan E et al (2010) In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong inter-individual variability. J Agric Food Chem 58:10236–10246
Macfarlane GT, Cummings JH, Macfarlane S, Gibson GR (1989) Influence of retention time on degradation of pancreatic-enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol 67:521–527
Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W (2009) How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10:1571–1581
Marzorati M, Vanhoecke B, De Ryck T, Sadaghian Sabadad M, Pinheiro I, Possemiers S et al (2014) The HMI module: a new in vitro tool to study the host microbiome interactions from the human gastrointestinal tract. BMC Microbiol 14:133
Miller TL, Wolin MJ (1981) Fermentation by the human large-intestine microbial community in an in vitro semicontinuous culture system. Appl Environ Microbiol 42:400–407
Minekus M, Smeets-Peeters M, Bernalier A, Marol-Bonnin S, Havenaar R, Marteau P et al (1999) A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl Microbiol Biotechnol 53:108–114
Molly K, Vandewoestijne M, Verstraete W (1993) Development of a 5-step multichamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol 39:254–258
Molly K, Vandewoestyne M, Desmet I, Verstraete W (1994) Validation of the simulator of the human intestinal microbial ecosystem (SHIME) reactor using microorganism-associated activities. Microb Ecol Health Dis 7:191–200
Peppercorn, Goldman (1972) The role of intestinal bacteria in the metabolism of salicyl azo-sulphapyridines. J Pharmacol Exp Ther 181:555–562
Possemiers S, Verthe K, Uyttendaele S, Verstraete W (2004) PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 49:495–507
Possemiers S, Bolca S, Grootaert C, Heyerick A, Decroos K, Dhooge W et al (2006) The prenylflavonoid isoxanthohumol from hops (Humulus lupulus L.) is activated into the potent phytoestrogen 8-prenylnaringenin in vitro and in the human intestine. J Nutr 136:1862–1867
Possemiers S, Pinheiro I, Verhelst A, Van den Abbeele P, Maignien L, Laukens D et al (2013) A dried yeast fermentate selectively modulates both the luminal and mucosal gut microbiota and protects against inflammation, as studied in an integrated in vitro approach. J Agric Food Chem 61:9380–9392
Sokol H, Pigneur B, Watterlot L et al (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736
Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gerard P et al (2010) Microbial community development in a dynamic gut model is reproducible, colon-region specific and selects for Bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol 76:5237–5246
Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S (2011) The host selects mucosal and luminal associations of co-evolved gut microbes: a novel concept. FEMS Microbiol Rev 35:681–704
Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O et al (2013) Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7:949–961
van Duynhoven J, Vaughan E, Jacobs D, Kemperman R, van Velzen E, Gross G et al (2011) The metabolic fate of polyphenols in the human superorganism. Proc Natl Acad Sci U S A 108:S4531–S4538
Vermeiren J, Van den Abbeele P, Laukens D, Visgnaes LK, De Vos M et al (2012) Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment. FEMS Microbiol Ecol 79:685–696
Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L et al (2009) Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis 15:653–660
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K et al (2009) Initial community evenness favours functionality under selective stress. Nature 458:623–626
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 The Author(s)
About this chapter
Cite this chapter
Van de Wiele, T., Van den Abbeele, P., Ossieur, W., Possemiers, S., Marzorati, M. (2015). The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In: Verhoeckx, K., et al. The Impact of Food Bioactives on Health. Springer, Cham. https://doi.org/10.1007/978-3-319-16104-4_27
Download citation
DOI: https://doi.org/10.1007/978-3-319-16104-4_27
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-15791-7
Online ISBN: 978-3-319-16104-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)