Abstract
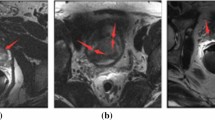
Automatic and accurate tumor segmentation on medical images is in high demand to assist physicians with diagnosis and treatment. However, it is difficult to obtain massive amounts of annotated training data required by the deep-learning models as the manual delineation process is often tedious and expertise required. Although self-supervised learning (SSL) scheme has been widely adopted to address this problem, most SSL methods focus only on global structure information, ignoring the key distinguishing features of tumor regions: local intensity variation and large size distribution. In this paper, we propose Scale-Aware Restoration (SAR), a SSL method for 3D tumor segmentation. Specifically, a novel proxy task, i.e. scale discrimination, is formulated to pre-train the 3D neural network combined with the self-restoration task. Thus, the pre-trained model learns multi-level local representations through multi-scale inputs. Moreover, an adversarial learning module is further introduced to learn modality invariant representations from multiple unlabeled source datasets. We demonstrate the effectiveness of our methods on two downstream tasks: i) Brain tumor segmentation, ii) Pancreas tumor segmentation. Compared with the state-of-the-art 3D SSL methods, our proposed approach can significantly improve the segmentation accuracy. Besides, we analyze its advantages from multiple perspectives such as data efficiency, performance, and convergence speed.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018)
Bilic, P., et al.: The liver tumor segmentation benchmark (lits). arXiv preprint arXiv:1901.04056 (2019)
Chen, L., Bentley, P., Mori, K., Misawa, K., Fujiwara, M., Rueckert, D.: Self-supervised learning for medical image analysis using image context restoration. Med. Image Anal. 58, 101539 (2019)
Doersch, C., Gupta, A., Efros, A.A.: Unsupervised visual representation learning by context prediction. In: IEEE International Conference on Computer Vision, pp. 1422–1430 (2015)
Feng, R., et al.: Parts2whole: self-supervised contrastive learning via reconstruction. In: Domain Adaptation and Representation Transfer, and Distributed and Collaborative Learning, pp. 85–95 (2020)
Gidaris, S., Singh, P., Komodakis, N.: Unsupervised representation learning by predicting image rotations. arXiv preprint arXiv: 1803.07728 (2018)
Haghighi, F., Taher, M.R.H., Zhou, Z., Gotway, M.B., Liang, J.: Learning semantics-enriched representation via self-discovery, self-classification, and self-restoration. In: Medical Image Computing and Computer Assisted Intervention, pp. 137–147 (2020)
Isensee, F., Jäger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: Automated design of deep learning methods for biomedical image segmentation. arXiv preprint arXiv:1904.08128 (2019)
Kingma, D., Ba, J.: Adam: a method for stochastic optimization. In: International Conference on Learning Representations, vol. 42 (2014)
Litjens, G.J.S., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Setio, A.A.A., Jacobs, C., Gelderblom, J., van Ginneken, B.: Automatic detection of large pulmonary solid nodules in thoracic CT images. Med. Phys. 42, 5642–5653 (2015)
Shin, H.C., et al.: Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imag. 35(5), 1285–1298 (2016)
Simpson, A.L., et al.: A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:1902.09063 (2019)
Taleb, A., et al.: 3D self-supervised methods for medical imaging. arXiv preprint arXiv:2006.03829 (2020)
Zhou, Z., Sodha, V., Pang, J., Gotway, M.B., Liang, J.: Models genesis. Med. Image Anal. 67, 101840 (2021)
Zhuang, X., Li, Y., Hu, Y., Ma, K., Yang, Y., Zheng, Y.: Self-supervised feature learning for 3D medical images by playing a rubik’s cube. In: Medical Image Computing and Computer Assisted Intervention, pp. 420–428 (2019)
Zongwei, Z., et al.: Models genesis: Generic autodidactic models for 3D medical image analysis. In: Medical Image Computing and Computer Assisted Intervention, pp. 384–393 (2019)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D u-net: learning dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer Assisted Intervention, pp. 424–432 (2016)
Acknowledgement
This work is supported partially by SHEITC (No. 2018-RGZN-02046), 111 plan (No. BP0719010), and STCSM (No. 18DZ2270700).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, X., Feng, S., Zhou, Y., Zhang, Y., Wang, Y. (2021). SAR: Scale-Aware Restoration Learning for 3D Tumor Segmentation. In: de Bruijne, M., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. MICCAI 2021. Lecture Notes in Computer Science(), vol 12902. Springer, Cham. https://doi.org/10.1007/978-3-030-87196-3_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-87196-3_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87195-6
Online ISBN: 978-3-030-87196-3
eBook Packages: Computer ScienceComputer Science (R0)