Abstract
Compound toxicity prediction is a very challenging and critical task in the drug discovery and design field. Traditionally, cell or animal-based experiments are required to confirm the acute oral toxicity of chemical compounds. However, these methods are often restricted by availability of experimental facilities, long experimentation time, and high cost. In this paper, we propose a novel convolutional neural network regression model, named BESTox, to predict the acute oral toxicity (\(LD_{50}\)) of chemical compounds. This model learns the compositional and chemical properties of compounds from their two-dimensional binary matrices. Each matrix encodes the occurrences of certain atom types, number of bonded hydrogens, atom charge, valence, ring, degree, aromaticity, chirality, and hybridization along the SMILES string of a given compound. In a benchmark experiment using a dataset of 7413 observations (train/test 5931/1482), BESTox achieved a squared correlation coefficient (\(R^2\)) of 0.619, root-mean-squared error (RMSE) of 0.603, and mean absolute error (MAE) of 0.433. Despite of the use of a shallow model architecture and simple molecular descriptors, our method performs comparably against two recently published models.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Measuring the chemical and physiological properties of chemical compounds are fundamental tasks in biomedical research and drug discovery [19]. The basic idea of modern drug design is to search chemical compounds with desired affinity, potency, and efficacy against the biological target that is relevant to the disease of interest. However, not only that there are tens of thousands known chemical compounds existed in nature, but many more artificial chemical compounds are being produced each year [9]. Thus, the modern drug discovery pipeline is focused on narrowing down the scope of the chemical space where good drug candidates are [7, 11]. Potential lead compounds will be subjected to further experimental validation on their pharmacodynamics and pharmacokinetic (PD/PK) properties [2, 14]; the latter includes absorption, distribution, metabolism, excretion, and toxicity (ADME/T) measurements. Traditionally, chemists and biologists conduct cell-based or animal-based experiments to measure the PD/PK properties of these compounds and their actual biological effects in vivo. However, these experiments are not only high cost in terms of both time and money, the experiments that involve animal testings are increasingly subjected to concerns from ethical perspectives [1].
Among all measured properties, toxicity of a compound is the most important one which must be confirmed before approval of the compound for medication purposes [16]. There are different ways to classify the toxicity of a compound. For example, based on systemic toxic effects, the common toxicity types include acute toxicity, sub-chronic toxicity, chronic toxicity, carcinogenicity developmental toxicity and genetic toxicity [22]. On the other hand, based on the toxicity effects area, toxicity can also be classified as hepatotoxicity, ototoxicity, ocular toxicity, etc. [15]. Therefore, there is a great demand for accurate, low-cost and time-saving toxicity prediction methods for different toxicity categories.
Toxicity of a chemical compound is associated with its chemical structure [17]. A good example is the chiral compounds. This kind of compounds and their isomers have highly similar structures but only slight differences in molecular geometry. Their differences cause them to possess different biological properties. For example, the drug Dopa is a compound for treating the Parkinson disease. The d-isomer form of this compound has severe toxicity whereas the l-isomer form does not [12]. Therefore, only its levorotatory form can be used for medical treatments. This property-structure relationship is often described as quantitative structure-activity relationship (QSAR) and have been widely used in the prediction of different properties of compounds [4, 24]. Based on the same idea, toxicities of a compound, being one of the most concerned properties, can be predicted via computational means as a way to select more promising candidates before undertaking further biological experiments.
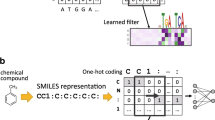
The Simplified Molecular Input Line Entry System, also called SMILES [20, 21], is a linear representation of a chemical compound. It is a short ASCII string describing the composition, connectivity, and charges of atoms in a compound. An example is shown in Fig. 1. The compound is called Morphine; it is originated from the opiate family and is found to exist naturally in many plants and animals. Morphine has been widely used as a medication to relief acute and chronic pain of patients. Nowadays, compounds are usually converted into their SMILES strings for the purpose of easy storage into databases or for other computational processing such as machine learning. Common molecular toolkits such as RDkit [8] and OpenBabel [13] can convert a SMILES string to its 2D and 3D structures, and vice versa.
In recent years, machine learning has become the mainstream technique in natural language processing (NLP). Among all machine learning applications for NLP, text classification is the most widely studied. Based on the input text sentences, a machine learning-based NLP model analyzes the organization of words and the types of words in order to categorize the given text. Two pioneering NLP methods are textCNN [6] and ConvNets [26]. The former method introduced a pretrained embedding layer to encode words of input sentences into fixed-size feature vectors with padding. Then, feature vectors of all words were combined to form a sentence matrix that was fed into a standard convolutional neural network (CNN) model. This work was considered a breakthrough at that time and accumulated over 5800 citations since 2014 (as per Google Scholar). Another spotlight paper in NLP for text classification is ConvNets [26]. Instead of analyzing words in a sentence, this model exploited simple one-hot encoding method at the character level for 70 unique characters in sentence analysis. The success of these methods in NLP shed lights to other applications that have only texts as raw data.
Compound toxicity prediction can be considered as a classification problem too. Recently, Hirohara et al. [3] proposed a new CNN model for toxicity classification based on character-level encoding. In this work, each SMILES character is encoded into a 42-dimensional feature vector. The CNN model based on this simple encoding method achieved an area-under-curve (AUC) value of 0.813 for classification of 12 endpoints using the TOX21 dataset [18]. The best AUC score in TOX21 challenge is 0.846 which is achieved by DeepTox [10]. Despite of its higher accuracy, the DeepTox model is extremely complex. It requires heavy feature engineering from a large pool of static and dynamic features derived from the compounds or indirectly via external tools. The classification model is ensemble-based combining deep neural network (DNN) with multiple layers of hidden nodes ranging from \(2^{10}\) to \(2^{14}\) nodes. The train dataset for this highly complex model was comprised of over 12,000 observations and superior predictive performance was demonstrated.
Besides classification, toxicity prediction can be seen as a regression problem when the compound toxicity level is of concern. Like other QSAR problems, toxicity regression is a highly challenging task due to limited data availability and noisiness of the data. With limited data, the use of simpler model architecture is preferred to avoid the model being badly overfitted. In this work, we have focused on the regression of acute oral toxicity of chemical compounds. Two recent works [5, 23] were found to solve this problem where the maximally achievable \(R^2\) is only 0.629 [5].
2 Materials and Methods
2.1 Dataset
In this study, we developed a regression model for acute oral toxicity prediction. The prediction task is to estimate the median lethal dose, \(LD_{50}\), of the compound; this is the dose required to kill half the members of the tested population. A small \(LD_{50}\) value indicates high toxicity level whereas a large \(LD_{50}\) value indicates low toxicity level of the compound. Based on the \(LD_{50}\) value, compounds can be categorized into four levels as defined by the United States Environmental Protection Agency (EPA) (see Table 1).
The rat acute oral toxicity dataset used in this study was kindly provided by the author of TopTox [23]. This dataset was also used in the recent study of computational toxicity prediction by Karim et al. [5]. For \(LD_{50}\) prediction task, the dataset contains 7413 samples; out of which 5931 samples are for training and 1482 samples are for testing. The original train/test split was deliberately made to maintain similar distribution of the train and test datasets to facilitate learning and model validation. It is noteworthy that as the actual \(LD_{50}\) values were in a wide range (train set: 0.042 mg/kg to 99947.567 mg/kg, test set: 0.020 mg/kg to 114062.725 mg/kg), the \(LD_{50}\) values were first transformed to mol/kg format, and then scaled logarithmically to \(-log_{10}\left( LD_{50} \right) \). Finally, the processed experimental values range from 0.470 to 7.100 in the train set and 0.291 to 7.207 in the test set.
2.2 Binary Encoding Method for SMILES (BES)
As a SMILES string is not an understandable input format for general machine learning methods, it needs to be converted or encoded into a series of numerical values. Ideally, these values should capture the characteristics of the compound and correlates to the interested observables. The most popular way to encode a SMILE is to use molecular fingerprints such as Molecular Access System (MACCS) and extended connectivity fingerprint (ECFP). However, fingerprint algorithms generate high dimensional and sparse matrices which make learning difficult.
Here, in order to solve the regression task for oral toxicity prediction. Inspired by the work of Hirohara et al. [3], we proposed the modified Binary Encoding method for SMILES, named BES for short. In BES, each character is encoded by a binary vector of 56 bits. Among them 26 bits are for encoding the SMILES alphabets and symbols by the one-hot encoding approach; 30 bits are for encoding various atomic properties including number of bonded hydrogens, formal charge, valence, ring atom, degree, aromaticity, chirality, and hybridization. The feature types and corresponding size of the feature is listed in Table 2.
As the maximum length of SMILES strings in our dataset is 300, the size of the feature matrix for one SMILES string was defined to be \(56\times 300\). For a SMILES string that is shorter than 300 in length, zero padding was applied. Figure 2 illustrates how BES works.
2.3 Model Architecture
Our prediction model is a conventional CNN model with convolutional layers to extract features, pooling layers to reduce dimensionality of the feature matrix and to prevent overfitting, and a multi-layer neural network to correlate features to \(LD_{50}\) values. To decide the model architecture and to tune hyperparameters of the model, a grid search method was employed. Table 3 shows the hyperparameters and their ranges of values within which the model was optimized. In each grid search process, the model training was run for 500 epochs and the mean-squared error (MSE) loss of the model in 5-fold cross validation was used as a criteria for model selection. The optimal parameters are also presented in Table 3. The final production model was trained using the optimal parameters and the entire train dataset. The maximum training epoch was 1000; early stop method was used to prevent the problem of overfitting.
MSE loss of training BESTox model with optimal hyperparameters in Table 3.
The architecture of our optimized CNN model is presented in Fig. 3. The model contains two convolutional layers (Conv) with 512 and 1024 filters respectively. After each convolutional layer is an average pooling layer and a batch normalization layer (BN). Then, a max pooling layer is used before the learned features fed into the fully connected layers (FC). Four FCs containing 2048, 1024, 512, and 256 hidden nodes were found to be the optimal combination for toxicity prediction and the ReLU function is used to generate the prediction output.
2.4 Implementation
All implementations were done using Python 3.6.9 with the following libraries: Anaconda 4.7.0, RDKit v2019.09.2.0, Pytorch 1.2.0 and CUDA 10.0. We used GetTotalNumHs, GetFormalCharge, GetChiralTag, GetTotalDegree, IsInRing, GetIsAromatic, GetTotalValence and GetHybridization functions from RDkit to calculate atom properties. Our model was trained and tested in a workstation equipped with two NVIDIA Tesla P100 GPUs.
3 Results
Training of the final production model was performed using the optimal parameters obtained from the result of our extensive grid search. Figure 4 shows the evolution of MSE over the number of training cycles. The training stopped at the 900-th epoch with MSE of 0.016. Table 4 shows the performances of our model in the train and test sets. The training performance is excellent which gives R\(^2\) of 0.982 as all the data was used to construct the model. For the test set, the model predicts with R\(^2\) of 0.619, RMSE of 0.603, and MAE of 0.433.
Figure 5 shows the scatterplot of BESTox prediction on the test data. We can see that prediction is better for compounds with lower toxicity (lower \(-log_{10}\left( LD_{50} \right) \)) and worse for those with higher toxicity. This may be due to fewer data available in the train set for higher toxicity compounds. Thus, we also tested our model on samples with target values less than 3.5 in the test set (1255 samples out of total 1482 samples, the sample coverage is more than 84%). In this case, the performance of our model is improved: RMSE is decreased from 0.603 to 0.516 and MAE is reduced from 0.433 to 0.385.
Table 5 presents the comparative performance of BESTox to two existing acute oral toxicity prediction models, the ST-DNN model from TopTox and the DT+SNN model from Karim et al. [5]. Results show that our model is slightly better than ST-DNN with respect to \(R^2\) and MAE. The best performed model is DT+SNN which has a correlation of 0.629; but RMSE and MAE were not provided in the original study.
The closeness of the performance metrics of BESTox to two existing models suggest that our model performs on par with them. Nevertheless, it should be mentioned that while our model has employed simple features and relatively simple model architecture, ST-DNN and DT+SNN relied on highly engineered input features and complex ensemble-based model architectures. For ST-DNN [23], they combined 700 element specific topological descriptors (ESTD) and 330 auxiliary descriptors as candidates to generate the feature vectors for prediction (our model uses only 56 features). In addition, their model included ensemble of two different types of classifiers, namely, deep neural network (DNN) and gradient boosted decision tree (GBDT). Combining predictions from several classifiers is an easy way to improve prediction accuracy, however, the complexity introduced into the model makes the already "black box model" more difficult to understand. For the recent DT+SNN model [5], they used decision trees (DT) to select 817 different descriptors generated from the PaDEL tools [25]. Although their shallow neural network (SNN) architecture required short model training time, more time was spent on feature generation and selection. Different combination of features were used depending on the tasks to be predicted, which had high computational cost. Here, BESTox has achieved results comparable to these more complex models with simple binary features and model architecture, showing the power of our method.
4 Conclusion
In this paper, we present our new method BESTox for acute oral toxicity prediction. Inspired by NLP techniques for text classification, we have designed a simple character-level encoding method for SMILES called the binary-encoded SMILES (BES). We have developed a shallow CNN to learn the BES matrices to predict the \(LD_{50}\) values of compounds. We trained our model on the rat acute oral toxicity data, tested and compared to two other existing models. Despite the simplicity of our method, BESTox has achieved a good performance with \(R^2\) of 0.619, comparable to the single-task model proposed by TopTox [23] but slightly inferior to the hybrid decision tree and shallow neural network model by Karim et al. [5].
Future improvement of BESTox will be focused on extending the scope of datasets. As shown in the work of Wu et al. [23], multitask learning can improve performance of prediction models due to availability of more data on different toxicity effects. The idea of multitask technique is to train a model with multiple training sets; each set corresponds to one toxicity prediction task. Feeding the learners with different toxicity data helps them to learn common latent features of molecules offered by different datasets.
References
Bailey, J., Balls, M.: Recent efforts to elucidate the scientific validity of animal-based drug tests by the pharmaceutical industry, pro-testing lobby groups, and animal welfare organisations. BMC Med. Ethics 20, 16 (2019)
Dean, A., Lewis, S.: Screening: Methods for Experimentation in Industry, Drug Discovery, and Genetics. Springer, Cham (2006). https://doi.org/10.1007/0-387-28014-6
Hirohara, M., Saito, Y., Koda, Y., Sato, K., Sakakibara, Y.: Convolutional neural network based on SMILES representation of compounds for detecting chemical motif. BMC Bioinform. 19, 526 (2018)
Idakwo, G., et al.: A review on machine learning methods for in silico toxicity prediction. J. Environ. Sci. Health Part C 36(4), 169–191 (2018)
Karim, A., Mishra, A., Newton, M.H., Sattar, A.: Efficient toxicity prediction via simple features using shallow neural networks and decision trees. ACS Omega 4(1), 1874–1888 (2019)
Kim, Y.: Convolutional neural networks for sentence classification. arXiv preprint arXiv:1408.5882 (2014)
Kubinyi, H., Mannhold, R., Timmerman, H.: Virtual Screening for Bioactive Molecules, vol. 10. Wiley, Hoboken (2008)
Landrum, G., et al.: RDkit: open-source cheminformatics (2006)
Llanos, E.J., Leal, W., Luu, D.H., Jost, J., Stadler, P.F., Restrepo, G.: Exploration of the chemical space and its three historical regimes. Proc. Natl. Acad. Sci. 116(26), 12660–12665 (2019)
Mayr, A., Klambauer, G., Unterthiner, T., Hochreiter, S.: DeepTox: toxicity prediction using deep learning. Front. Environ. Sci. 3, 80 (2016)
McInnes, C.: Virtual screening strategies in drug discovery. Curr. Opin. Chem. Biol. 11(5), 494–502 (2007)
Nguyen, L.A., He, H., Pham-Huy, C.: Chiral drugs: an overview. Int. J. Biomed. Sci. IJBS 2(2), 85 (2006)
O’Boyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., Hutchison, G.R.: Open Babel: an open chemical toolbox. J. Cheminform. 3(1), 33 (2011)
Oprea, T.I., Matter, H.: Integrating virtual screening in lead discovery. Curr. Opin. Chem. Biol. 8(4), 349–358 (2004)
Quintanilha, J.C.F., Berlofa, M.: New promising approaches to treatment of chemotherapy-induced toxicities. AvidScience Chemother. 2–52 (2017)
Raies, A.B., Bajic, V.B.: In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 6(2), 147–172 (2016)
Roy, K., Kar, S., Das, R.: Chapter 7–validation of QSAR models. Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment, pp. 231–289 (2015)
Tice, R.R., Austin, C.P., Kavlock, R.J., Bucher, J.R.: Improving the human hazard characterization of chemicals: a TOX21 update. Environ. Health Perspect. 121(7), 756–765 (2013)
Ting, N.: Dose Finding in Drug Development. Springer, Cham (2006). https://doi.org/10.1007/0-387-33706-7
Weininger, D.: SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28(1), 31–36 (1988)
Weininger, D., Weininger, A., Weininger, J.L.: SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 29(2), 97–101 (1989)
Wexler, P., Gad, S.C., et al.: Encyclopedia of Toxicology. Academic Press, Cambridge (1998)
Wu, K., Wei, G.W.: Quantitative toxicity prediction using topology based multitask deep neural networks. J. Chem. Inf. Model. 58(2), 520–531 (2018)
Wu, Y., Wang, G.: Machine learning based toxicity prediction: from chemical structural description to transcriptome analysis. Int. J. Mol. Sci. 19(8), 2358 (2018)
Yap, C.W.: Padel-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32(7), 1466–1474 (2011)
Zhang, X., Zhao, J., LeCun, Y.: Character-level convolutional networks for text classification. In: Advances in Neural Information Processing Systems, pp. 649–657 (2015)
Acknowledgments
This work was supported by University of Macau (Grant no. MYRG2017-00146-FST).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Chen, J., Cheong, HH., Siu, S.W.I. (2020). BESTox: A Convolutional Neural Network Regression Model Based on Binary-Encoded SMILES for Acute Oral Toxicity Prediction of Chemical Compounds. In: Martín-Vide, C., Vega-Rodríguez, M., Wheeler, T. (eds) Algorithms for Computational Biology. AlCoB 2020. Lecture Notes in Computer Science(), vol 12099. Springer, Cham. https://doi.org/10.1007/978-3-030-42266-0_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-42266-0_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-42265-3
Online ISBN: 978-3-030-42266-0
eBook Packages: Computer ScienceComputer Science (R0)