Abstract
Diffraction-limited lens-based optical microscopy fails to discern fluorescent features closer than \(\sim \)200 nm. All super-resolution microscopy (nanoscopy) approaches that fundamentally overcome the diffraction barrier rely on fluorophores that can adopt different states, typically a fluorescent ‘on-’state and a dark, non-fluorescent ‘off-’state. In reversible saturable optical linear fluorescence transitions (RESOLFT) nanoscopy, light is applied to induce transitions between two states and to switch fluorophores on and off at defined spatial coordinates. RESOLFT nanoscopy relies on metastable reversibly switchable fluorophores. Thereby, it is particularly suited for live-cell imaging, because it requires relatively low light levels to overcome the diffraction barrier. Most implementations of RESOLFT nanoscopy utilize reversibly photoswitchable fluorescent proteins (RSFPs), which are derivatives of proteins from the green fluorescent protein (GFP) family. In recent years, analysis of the molecular mechanisms of the switching processes have paved the way to a rational design of new RSFPs with superior characteristics for super-resolution microscopy. In this chapter, we focus on the newly developed RSFPs, the light-driven switching mechanisms and the use of RSFPs for RESOLFT nanoscopy.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Overcoming the Diffraction Barrier
Optical fluorescence microscopy allows to discern protein distributions in cells. Over the last century, cell biology experienced countless discoveries based on the microscopic visualization of (sub-)cellular structures and their dynamics. Eukaryotic cells, which generally have a diameter of 10–300 \(\upmu \)m, are relatively small objects crowded with proteins. Hence, conventional optical microscopy, whose resolution is fundamentally limited by diffraction to about 200 nm, inescapably faces the fact that the finest details are not resolvable.
Resolution in microscopy means the separation of distinct features, such as fluorescently labelled proteins. In conventional diffraction limited microscopy, such as widefield (epifluorescence) or confocal laser scanning microscopy, all fluorophores in closer proximity to each other than the diffraction limit are excited together, they emit together, and their emissions diffract together and therefore they are detected together [1]. Hence, with these approaches, structures closer than the diffraction limit are inseparable. The key to fundamentally overcome the diffraction barrier is to render adjacent molecules discernible for a short period of time, preventing different molecules within the same diffraction region from being detected together [2]. This separation can be implemented either in a coordinate-targeted or in a coordinate-stochastic way (for reviews see [1, 3]).
In coordinate-targeted super-resolution microscopy techniques such as RESOLFT nanoscopy, a light pattern is used to induce transitions between two states and to switch fluorophores on and off at defined spatial coordinates. In the simplest scenario, a single beam creating one intensity minimum in a doughnut-shaped pattern is used, but also approaches with several or many minima are possible. At these minima (zeros), there is no off-switching, and the fluorophores in the on-state can fluoresce. These light patterns are scanned over the sample, in order to record a full image. Such scanning approaches require multiple on-off-cycles of the fluorophores.
2 RSFPs for Live-Cell RESOLFT Nanoscopy
In its initial definition, the term ‘RESOLFT nanoscopy’ covered all coordinate-targeted nanoscopy approaches relying on two distinct (fluorophore) states including STED nanoscopy [4]. Later, this term was primarily used for coordinate-targeted nanoscopy approaches that rely on metastable fluorophores, most prominently on reversibly (photo-)switchable fluorescent proteins (RSFPs). Compared to other super-resolution microscopy techniques that overcome the diffraction barrier, RESOLFT nanoscopy requires remarkably low light dose to achieve nanoscale resolution. The light intensities used are similar to those applied in live-cell confocal fluorescence microscopy and up to six orders of magnitude lower than those in STED-microscopy. The total light dose deposited on the sample is lower by 3–4 orders of magnitude compared to coordinate-stochastic nanoscopy [5, 6]. As the light intensity is an important factor that determines phototoxicity [7], RESOLFT nanoscopy is particularly suitable for live-cell imaging approaches.
RESOLFT microscopy relies on fluorophores that can be reversibly photoswitched between two metastable states. In the overwhelming majority of applications, these have been reversibly photoswitchable fluorescent proteins. RSFPs are structurally highly similar to the green fluorescent protein (GFP) . All GFP-based fluorescent proteins share the same overall structure of 11 \(\beta \)-sheets, forming a \(\beta \)-barrel with an \(\alpha \)-helix running through the center. The chromophore is autocatalytically formed out of three amino acids within the \(\alpha \)-helix requiring only oxygen as an external cofactor for its maturation (Fig. 9.1) [8]. In GFP, the chromophore consists of a hydroxyphenyl ring and an imidazolinone ring connected by a methine bridge. By rotation around the methine bridge, the chromophore can adopt either a cis or a trans conformation. The hydroxyphenyl ring can be protonated or deprotonated, thereby shifting the absorption spectrum by about 80–120 nm. The \(\beta \)-barrel shields the chromophore in its center, while numerous noncovalent bonds to surrounding amino acid residues and internal water molecules determine the chromophore position within the protein and its protonation state. The spectral properties of a specific RSFP are a result of the chromophore structure as well as of its interactions with the surrounding residues; therefore, those positions are key targets for mutagenesis to modify the properties of RSFPs.
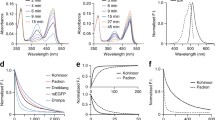
Structure of EGFP and its intrinsic chromophore. a Side view of the GFP barrel. b Top view of the GFP barrel with the internal chromophore in its center (PDB: 2Y0G). c Structure of the GFP chromophore. The chromophore consists of an imidazolinone and a hydroxyphenyl ring connected by a methine bridge
Schematic overview of the different switching mechanisms. a Negative switching RSFPs can be switched off with light of the wavelength used for fluorescence excitation. b Positive switching RSFPs are switched on by the fluorescence excitation light. c Proteins with decoupled switching are excited with light of a different wavelength than used for on- or off-switching. For each switching mechanism, schematic chromophore structures in the on- and the off-state, examples for the switching wavelengths, and the respective absorption and emission spectra are shown
3 Photoswitching Mechanisms of RSFPs
Conventional RSFPs can be classified according to their switching mode, i.e. a ‘positive’ or a ‘negative’ switching mode (Fig. 9.2) [9]. In negative switching RSFPs, the same wavelength that induces fluorescence also switches the RSFP from the on- to the off-state. In contrast, in positive switching RSFPs, the light that induces fluorescence also transfers the protein from the off- to the on-state. Thereby, in conventional RSFPs, switching and fluorescence excitation are directly interconnected. The mechanistic principles of switching have been initially revealed by spectroscopy and crystallography of the RSFPs asFP595 and Dronpa in their respective on- and off-states [10, 11]. In recent years, numerous further studies [12,13,14,15,16], including detailed molecular dynamics studies, ultrafast spectroscopy and crystallography have led to impressive insights into the details of the light driven switching mechanism [17,18,19,20]. The mechanistic key event of the switching of all conventional RSFPs analyzed so far is a light induced cis-trans isomerization of the chromophore often combined with a protonation change [13]. This isomerization can be accompanied by shifts of the planarity of the chromophore and/or conformational changes of the chromophore pocket and modifications of the hydrogen-bonding network around the chromophore, influencing the chromophores’ ability to fluoresce. Almost all reported crystallographic structures of conventional RSFPs feature a trans-conformation of the chromophore in the off-state and a cis-conformation in the on-state [21]. However, it should be noted, while the majority of fluorescent proteins contain a chromophore in the cis-conformation, bright fluorescent proteins containing a chromophore in the trans-state exist [22]. The only reported RSFP so far with a different molecular switching mechanism is the yellow fluorescent RSFP Dreiklang [23] and its descendant Spoon [24] (Fig. 9.2c). In Dreiklang, fluorescence excitation is decoupled from switching, i.e. one wavelength is used for fluorescence excitation (515 nm), another for switching on (405 nm), and a third for switching off (355 nm). In Dreiklang, a light-driven reversible hydration/dehydration of the chromophoric imidazolinone ring causes switching by reversibly disrupting the \(\pi \)-conjugated electron system.
In the following, we will take a closer look at the different switching modes and their application in RESOLFT imaging.
3.1 Negative Switching Mode
Negative switching RSFPs are most commonly used for RESOLFT nanoscopy. In the dark state, the chromophore is found in the protonated trans-conformation, while in the on-state the chromophore is deprotonated in the cis-conformation (Fig. 9.2a). Excitation of the on-state chromophore results in fluorescence (quantum yield typically 0.1–0.9), switching (quantum efficiency typically in the range of 0.01) [25], or to other non-radiative processes. In comparison, the quantum efficiency for switching the protein from the off- to the on-state is generally substantially higher (quantum efficiency typically in the range of 0.1). The wavelength used for switching the RSFP from the off- into the on-state is usually 80–120 nm shorter than the wavelength for switching off and fluorescence excitation. The precise sequence of the intra-molecular events during the switching process, particularly the sequence of isomerization and protonation change, have been controversially discussed [21]. Recent results for Dronpa and IrisFP conclusively suggested that during the off-to-on transition the chromophore isomerization precedes the protonation change [20, 26].
In single-beam scanning RESOLFT nanoscopy, negative switching RSFPs are generally utilized according to the following imaging scheme: First, the proteins are switched to the on-state with a Gaussian beam, then the proteins in the periphery are switched off with a doughnut-shaped beam, and subsequently the remaining fluorescence in the center (whose size is smaller than the diffraction limit) is read out. The on-switching process has generally a higher quantum efficiency than the off-switching process. Thus, the off-switching step is the most time-consuming step, although fast switching proteins have been developed that reduced the dwell time strongly [5]. However, a too high quantum efficiency for switching off is also not desirable, as switching competes with fluorescence excitation in case of RSFPs with a negative switching mode.
3.2 Positive Switching Mode
All reported RSFPs with positive switching characteristics, including Padron [9], rsCherry [27] or asFP595 [28], share an Anthozoa heritage. In the off-state, the chromophore of the positive switching RSFPs is generally in the trans-state and deprotonated (Fig. 9.2b). Excitation into the absorption band of the deprotonated chromophore induces chromophore isomerization to the cis-state. In the cis-state, the protein is in an equilibrium between the protonated and the deprotonated state. Excitation of the protonated cis-chromophore switches the protein to the trans off-state, while excitation of the deprotonated cis-chromophore excites fluorescence. The protonation equilibrium of the chromophore in the cis- and the trans-states is immediately influenced by the amino acids forming the chromophore pocket. Reportedly, the exchange of few amino acids can induce a change in the switching mode of the respective fluorescent protein [9, 12, 27].
Using positive switching RSFPs, the optical system for beam scanning RESOLFT nanoscopy can be reduced to a Gaussian beam for switching on and fluorescence excitation and a doughnut-shaped beam for switching the protein off [29]. A potential advantage of RESOLFT imaging with positive switching fluorescent proteins is that the number of emitted photons during readout is not limited by a competing off-switching process.
3.3 Decoupled Switching Mode
The switching of Dreiklang and its descendants differs from the switching of conventional RSFPs by the fact that the excitation of fluorescence does not induce substantial switching [23]. The chromophore of the on-state protein is in an equilibrium between a protonated and a deprotonated state (Fig. 9.2c). Excitation of the deprotonated on-state chromophore at 515 nm results in fluorescence, while excitation of the protonated chromophore at 405 nm leads to a hydration of the C65 atom of the imidazolinone ring, thereby switching the protein to the off-state. More specifically, upon excitation into the absorption band of the protonated chromophore, an ultrafast excited state proton transfer occurs, presumably leading to a charge transfer to the imidizanolinone ring, which subsequently is protonated by Glu222, catalyzing the addition of a water molecule [30]. By this water addition, the \(\pi \)-conjugated electron system is shortened, resulting in the emergence of an absorption band at 395 nm. Excitation of this band results in a water elimination reaction, thereby converting the protein back to the on-state. The positioning of the water molecule reacting with the chromophore and the protonation state of the chromophore are crucial for the efficiency of this switching mechanism. This process is dependent on at least three key amino acids (Gly65, Tyr203, Glu222) [23].
The decoupling of switching from fluorescence excitation enables full control of the state of the protein, which is desirable in RESOLFT nanoscopy. Dreiklang has been used for RESOLFT nanoscopy [23, 31], although it is outperformed by other RSFPs, because some of its other properties (see below) are less favorable for this imaging modality.
4 RSFP Properties Important for RESOLFT Nanoscopy
The usefulness of a specific RSFP for live-cell RESOLFT nanoscopy depends on several factors. On the one hand, any usable RSFP must be a suitable fusion tag with a negligible dimerization tendency and a fast maturation rate. On the other hand, it needs to exhibit a combination of favorable photophysical characteristics. The four most important parameters for RESOLFT imaging are the brightness of the protein in its on-state, its switching speed, the residual fluorescence background in the ensemble off-state, and the switching fatigue (Fig. 9.3). These parameters, all of which can be influenced by mutagenesis, are discussed in detail in the following.
Important switching parameters. Exemplary switching curve of negative switching RSFPs when switched consecutively on and off with light of different wavelengths. Plotted is the emitted fluorescence intensity. Four key parameters can be determined from these switching curves. a effective brightness of the RSFP in the on-state. b Switching speed. c Residual fluorescence in the off-state. d Switching fatigue
4.1 Brightness
For a conventional fluorophore, the molecular brightness is defined as the product of the extinction coefficient and the quantum yield. RSFPs can adopt two different states with different quantum yields and in most cases the two states exhibit distinct absorption spectra. Generally, the molecular brightness of the off-state is very small but not necessarily zero. Next to the molecular brightness of the protein in the on-state, two other parameters, namely the effective brightness (of an ensemble of fully matured RSFPs in solution) and the effective cellular brightness (of RSFPs in a living cell) are used to describe and compare RSFPs. In a switching curve (Fig. 9.3a), the effective brightness is measured as the fluorescence intensity of the protein ensemble when switched fully into the on-state. In absolute terms, it is the average molecular brightness of the on-switched protein solution. As a RSFP solution may adopt an equilibrium between the on- and the off-state [9, 27], the effective brightness can differ drastically from the brightness at equilibrium.
The effective cellular brightness of a specific RSFP is influenced by a number of factors, such as its maturation time, its turnover-rate, its expression rate, its usability as a tag, temperature, local pH and other difficult to pinpoint factors. As RSFPs are primarily utilized for in vivo imaging, the effective cellular brightness is a very crucial parameter when RSFPs are compared. The cellular brightness can vary strongly between different experimental settings [32, 33] and therefore the data from different studies are difficult to compare.
4.2 Ensemble Switching Speed
The switching speed of an ensemble of RSFPs is typically in the microsecond to second range, depending on the light intensity applied [5, 25, 34, 35]. Since RESOLFT is an ensemble method, the ensemble switching speed is a key parameter to describe and compare RSFPs. In a switching curve (Fig. 9.3), this parameter describes the time needed to switch the ensemble from the maximal fluorescence intensity to the minimal fluorescence intensity (from the on- to the off-state) or vice versa (Fig. 9.3b). The speed is dependent on the applied light intensities and wavelengths; applying higher light intensities results in faster switching. As switching is a one photon process in both directions [36, 37], the speed and the applied light intensity for the switching are largely linearly correlated until saturation of the process sets in. The ensemble switching speed is determined primarily by the quantum efficiency for switching, by (often ill-defined) intermediate states, and by a crosstalk between the on- and the off-switching processes. While high quantum efficiencies for switching are beneficial for RESOLFT imaging by shortening the time consuming switching step, the connection between switching and fluorescence readout limits the number of collected photons in a single switching cycle. For point-scanning RESOLFT in living cells, the switching times are generally below 1 ms [5, 6], while in parallelized RESOLFT schemes, switching times of tens of milliseconds are suitable [34, 38].
4.3 Residual Fluorescence in the Off-State
The residual fluorescence in the off-state describes the percentage of the fluorescence signal from the off-state compared to the fluorescence signal of the on-state (Fig. 9.3c). The switching contrast is defined as the ratio between the fluorescence signal of the on-state and the fluorescence signal of the off-state measured on an ensemble of proteins.
The switching contrast is determined by the crosstalk between on- and off-switching and the molecular brightness of the on- and the off-state. Furthermore, fast thermal relaxation of the switched protein to the respective equilibrium state, as well as the population of intermediate states, can affect the reachable switching contrast. RSFPs useful for RESOLFT nanoscopy exhibit a contrast higher than 10 (a residual fluorescence below 10%), although smaller values of the residual fluorescence have been reported and are beneficial [25, 39].
4.4 Switching Fatigue
With every full switching cycle of an ensemble of RSFPs, a fraction of the proteins is destroyed. This fraction is generally described as the switching fatigue (given as percentage of the effective brightness) (Fig. 9.3d). Presumably, the switching fatigue is mechanistically related to the photostability of a fluorescent protein, which describes its stability while the protein is maintained and excited in the fluorescent on-state.
A low switching fatigue is critical for the usability of RSFPs in RESOLFT microscopy [40]. Photobleaching is highly dependent on the light intensity, as different intensity regimes of photobleaching and nonlinear effects of increasing light intensities have been reported [32, 41]. Therefore, lower light intensities typically result in reduced switching fatigue [39]. Very photostable RSFPs can be switched thousands of times before they are bleached to 50% of their initial brightness [5, 42].
Brightness, switching speed, contrast and switching fatigue are strongly intertwined. The introduction of switching into a conventional fluorescent protein as well as the increase of the switching speed of an existing RSFP by mutagenesis generally lead to a reduction in the fluorescence quantum yield [5, 39, 43, 44]. Furthermore, for negative switching RSFPs a correlation of the switching fatigue with the off-switching speed has been reported [25].
5 Overview of RSFPs for RESOLFT Nanoscopy
In the last decade, a number of new RSFPs have been engineered using semi-rational design strategies guided by the crystal structures and insights into the switching mechanism. Thereby, either conventional fluorescent proteins were made switchable, or the characteristics of existing RSFPs were modified. The members of this growing family of RSFPs have been used for a number of applications [45], including intracellular protein tracking [46], photochromic FRET [47], optogenetics [48], optoacoustics [49], and several super-resolution microscopy techniques including coordinate-stochastic approaches [9], SOFI [50, 51], (protected) STED [52, 53] and RESOLFT/NL-SIM [39, 42].
For each application, distinct RSFP properties are crucial and RSFPs have been optimized and adapted for the specific demands. In the following, we provide an overview on the published RSFPs emitting in the green, yellow and red regimes of the visible spectrum, which were used in RESOLFT microscopy or have properties beneficial for RESOLFT imaging (Fig. 9.4, Table 9.1).
Fluorescence emission wavelength versus molecular brightness of RSFPs. Displayed are the molecular brightness of the on-state (normalized to EGFP) and the fluorescence emission wavelength maximum of selected RSFPs. The selection is not comprehensive. Note that only RSFPs with a (potential) usability for nanoscopy are presented
5.1 RSFPs Emitting in the Green
RSFPs emitting in the green have been engineered based on several well-characterized fluorescent proteins isolated from Hydrozoan and Anthozoan species. The switching wavelengths of these green fluorescent RSFPs are at \(\sim \)488 and \(\sim \)405 nm, corresponding to the absorption bands of the deprotonated and the protonated chromophore. The first experimental implementation of live-cell RESOLFT nanoscopy utilized the negative switching rsEGFP [42], which has been tailored for RESOLFT nanoscopy based on the Hydrozoan EGFP [60]. Derived from rsEGFP, a number of other RSFPs were engineered such as families of rsEGFPs [5, 56], rsGreens [25], and rsFolders [14], with further optimized expression or modified switching characteristics for specific applications.
The EGFP derived RSFPs show very good tagging capabilities, are true monomers, and exhibit fast to very fast switching kinetics. Especially the high resistance of rsEGFP2 to switching fatigue at high and low light intensities makes it the most widely used RSFP in RESOLFT imaging to date.
The very bright Anthozoa protein Dronpa, which is based on the oligomeric protein 22G from Pectiniidae spec., was the first RSFP to be used in live-cell imaging [46]. Dronpa exhibits comparatively slow switching and very high switching fatigue; Dronpa’s crystal structures of the on- and off-state were solved early, guiding the development of models for the switching mechanisms and strategies for semi-rational protein design [10, 44]. Based on Dronpa, several RSFPs with improved switching properties including rsFastLime, Dronpa-M159T [44] and Dronpa-3 [43] were developed. The increase in switching speed of these mutants was always accompanied by a reduction of their respective molecular brightness; still, Dronpa-M159T is well suited for point-scanning RESOLFT [61].
A single-residue exchange at the position 159 (Met159Tyr) is sufficient to reverse the switching mode of rsFastLime, i.e. this mutation converted the negative switching RSFP rsFastLime to the positive switching rsFastLime-M159Y. Additional mutations led to the engineering of the bright, positive switching RSFP Padron, exhibiting a high switching contrast [9]. Further mutagenesis changed the switching characteristics and photostability of Padron and led to the development of Kohinoor [29] opening the door to live-cell RESOLFT with positive switching RSFPs.
In recent years, several RSFPs have been engineered using photoconvertible fluorescent proteins as scaffolds. Conventional photoconvertible fluorescent proteins are irreversibly switched from one emission maximum (generally green) to another one (generally red). The mutation Phe173Ser was shown to introduce negative switching to the photoconvertible Anthozoa proteins Dendra2 [62] and mEos [63], resulting in the multiphotochromic RSFPs NijiFP [55] and (with additional mutations) mIrisFP [64]. NijiFP and mIrisFP are both photoconvertible and negatively switchable in the red and green form.
All reported green to red photoconvertible fluorescent proteins have a histidine at the first amino acid position of their chromophore, which is essential for their photoconvertibility [65]. Exchanging this histidine by another amino acid residue can transform a photoconvertible protein into a negative switching RSFP. This was reported for the Anthozoa protein mEos2 [66]. By exchanging the His for different amino acids (Cys, Glu, Phe, Leu, Met, Ser, partially in combination with the Phe173Ser mutation), a series of negative switching RSFPs (mGeos-X), with switching properties similar to those of various Dronpa variants, was produced [57]. Likewise, the His62Leu mutation of the protein mEos3.1 [67] (a true monomer and bright mEos2 descendant), resulted in the RSFP Skylan-NS [39], which exhibits negative switching characteristics with high brightness and excellent switching contrast.
Following the same strategy, exchange of the chromophoric His in the photoconvertible Anthozoa FP mMaple3 [68], in combination with Met168Ala (corresponding to Met159 in Dronpa), resulted in a series of negative switching RSFPs (GMars-X) displaying various switching kinetics beneficial for different RESOLFT applications, in particular for parallelized RESOLFT [35, 54, 69].
5.2 RSFPs Emitting in the Yellow
Based on the yellow fluorescent protein Citrine (a mutant of GFP) [70], Dreiklang [23] and its descendant Spoon [24] were developed. As detailed above, in Dreiklang the switching is decoupled from fluorescence readout and this RSFP has been used for single-beam and parallelized RESOLFT imaging [23, 31, 38]. A limitation of Dreiklang is the comparatively low switching speed, the requirement for two UV light wavelengths for switching and its pronounced switching fatigue. However, in principle, the decoupled switching mechanism should be very beneficial for most RESOLFT applications. Remarkably, no conventional RSFPs have been reported based on a yellow fluorescent protein so far.
5.3 RSFPs Emitting in the Red
Even though the positive switching red RSFP asFP595 was the first RSFP used for proof of concept RESOLFT imaging with purified protein solutions [71], its obligate tetramerization and poor performance as a fusion tag hindered its use in live-cell imaging.
Derived from the Anthozoa protein mCherry, the RSFPs rsCherry, rsCherryRev [27] and rsCherryRev1.4 [59] were established. The high photostability and fast switching of rsCherryRev1.4 facilitated point-scanning and parallelized live-cell RESOLFT nanoscopy with this red emitting RSFP [38, 59], but its poor expression in mammalian cells and a tendency for dimerization limit its applicability.
Based on TagRFP [72], the red emitting RSFP rsTagRFP [47] was developed. It shows good contrast and strong changes in the absorption spectra of the on- and off-states. However, a low extinction coefficient and high switching fatigue prevented RESOLFT imaging in living cells. Only recently, a new family of red fluorescent RSFPs based on FusionRed was engineered [34]. The parent protein shows excellent tagging performance and rsFusionRed2 as well as rsFusionRed3 were successfully used in a parallelized RESOLFT approach [34].
In conclusion, a growing family of RSFPs for RESOLFT nanoscopy is available, each member having distinct properties. As many properties of RSFPs are depending on the light intensities used and the (cellular) environment, data recorded in different laboratories may be difficult to compare. Currently, outside the green fluorescence spectral regime, only a few RSFPs are available, underscoring the need for further protein engineering of red and infrared RSFPs.
6 Applications of RESOLFT Nanoscopy
Since the first proof of concept demonstration of RESOLFT nanoscopy using purified asFP595 [71], numerous RESOLFT live-cell applications have been reported [45]. It has been used in point-scanning (Fig. 9.5a) [5, 6, 14, 23, 25, 42, 73] as well as in various parallelized implementations (Fig. 9.5b) [34, 35, 38, 54, 56, 69, 74, 75].
Examples of RESOLFT imaging of living cells. a Point-scanning RESOLFT microscopy of intermediate filaments. Keratin19-rsEGFP2 expressed in living Ptk2 cells imaged in the confocal (left) and point-scanning RESOLFT mode (middle) [5]. The graphs show fluorescence intensity line profiles taken at sites indicated in the magnification (right). b Parallelized RESOLFT microscopy of intermediate filaments. Keratin19-rsEGFP-N205S expressed in living Ptk2 cells [56]. c Two color RESOLFT imaging. Vimentin-rsCherryRev1.4 and Keratin-rsEGFP2 were imaged in HeLa cells [59]. d Parallelized RESOLFT microscopy using a red RSFP. Living U2OS cells expressing rsFusionRed3-Vimentin [34]. e In vivo RESOLFT microscopy. Time-lapse point-scanning RESOLFT imaging of rsEGFP2-Tubulin facilitates the observation of tubulin dynamics in intact living fly larvae [6]. Images are adapted with permissions from the listed references. Scale bars: a 1 \(\upmu \)m; b 10 \(\upmu \)m; c 0.25 \(\upmu \)m; d 5 \(\upmu \)m; e 1 \(\upmu \)m
Arguably, rsEGFP2 is the most suited protein in the point-scanning mode, as it displays a combination of favorable properties, most prominently its strong resistance against switching fatigue. Amongst others, rsEGFP2 has been used for imaging of fixed cells that had been labeled by a primary antibody against a protein of interest that was detected by a fusion protein consisting of the IgG binding Z-domain of protein A and rsEGFP2 [73]. Live-cell RESOLFT imaging was established on CRISPR/Cas9 edited cell lines expressing rsEGFP2 fusion proteins at endogenous levels [76]. Likewise, rsEGFP2 was used for RESOLFT nanoscopy in living Drosophila melanogaster tissues and even in intact transgenic second instar fly larvae [6] (Fig. 9.5e). Using rsEGFP2, tens of images with a resolution below 50 nm can be taken for small fields of view with a frame rate of a few seconds. To improve specific characteristics, several variants such as rsGreen and rsFolder were generated and used for RESOLFT nanoscopy [14, 25]. The fast switching Dronpa variant Dronpa-M159T has similar switching kinetics as rsEGFP2, and it has also been used in a few RESOLFT applications [61, 77]. Next to these negative-switching RSFPs, the positive-switching RSFP Kohinoor [29] and the decoupled RSFP Dreiklang were used for cellular RESOLFT imaging [23, 31, 38].
Several strategies have been applied to perform dual color live-cell RESOLFT nanoscopy, including an approach that exploited the different switching speeds and fluorescence lifetimes of two RSFPs [78]. Despite the fact that the red fluorescent proteins are outperformed by the green ones, some studies utilized a red fluorescent RSFP in combination with a green [59] or yellow fluorescent RSFP [38] for dual label RESOLFT imaging (Fig. 9.5c).
Because of the rather long dwell times, single beam scanning RESOLFT nanoscopy of large fields of view is rather slow and recording of a single image may take several tens of seconds. To address this limitation, several parallelization strategies have been successfully implemented. The first realization of parallelized RESOLFT relied on incoherently superimposed orthogonal standing light waves that allowed to image whole cells in a few seconds [56], and other strategies were reported subsequently (Fig. 9.5b, d) [38, 74, 79]. RESOLFT nanoscopy was also implemented in a light sheet approach [80] and has been combined with STED nanoscopy [52].
6.1 Other Fluorophores for RESOLFT Nanoscopy
Most current realizations of RESOLFT imaging used RSFPs from the GFP family. Only recently, several other photochromic fluorophores have been utilized for RESOLFT-type nanoscopy. This includes rsLov1, an engineered reversibly switchable protein based on the bacterial photoreceptor YtvA from Bacillus subtilis that binds flavin mononucleotide as a chromophore [81]. Also, organic fluorophores were successfully used, including carboxylated photoswitchable diarylethenes [82] and Cy3-Alexa647 heterodimers [83]. Currently, however, these novel probes are outperformed by RSFPs based on the GFP backbone, with regard to basically all photophysical parameters important for live-cell RESOLFT nanoscopy.
7 Outlook
RESOLFT nanoscopy opens up very flexible imaging schemes. For example, the attained resolution can be adjusted so that the resolution is decreased in favor of reduced phototoxicity, or of an increased recording speed, or in favor of the number of images taken before bleaching. This flexibility, in combination with the relatively low light intensities required, makes RESOLFT nanoscopy an exciting option for live-cell applications.
Recent developments enabling a parallelization of the method allowed to overcome the rather low imaging speed of single-beam solutions. Still, the potential of RESOLFT nanoscopy has not been fully exploited as yet. This is mainly due to the lack of suitable RESOLFT probes emitting in the red and infrared spectral regime, which hinders the development of robust multi-color RESOLFT applications. We predict that this gap will be closed in the near future, as a number of promising templates for the development of such probes are emerging [84].
References
Sahl, S.J., Hell, S.W., Jakobs, S.: Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18(11), 685–701 (2017)
Hell, S.W.: Toward fluorescence nanoscopy. Nat. Biotechnol. 21(11), 1347–1355 (2003)
Sigal, Y.M., Zhou, R., Zhuang, X.: Visualizing and discovering cellular structures with super-resolution microscopy. Science 361(6405), 880–887 (2018)
Hell, S.W., Dyba, M., Jakobs, S.: Concepts for nanoscale resolution in fluorescence microscopy. Curr. Opin. Neurobiol. 14(5), 599–609 (2004)
Grotjohann, T., Testa, I., Reuss, M., Brakemann, T., Eggeling, C., Hell, S.W., Jakobs, S.: rsEGFP2 enables fast RESOLFT nanoscopy of living cells. Elife 1, e00248 (2012)
Schnorrenberg, S., Grotjohann, T., Vorbruggen, G., Herzig, A., Hell, S.W., Jakobs, S.: In vivo super-resolution RESOLFT microscopy of drosophila melanogaster. Elife 5 (2016)
Waldchen, S., Lehmann, J., Klein, T., van de Linde, S., Sauer, M.: Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 5, 15348 (2015)
Tsien, R.Y.: The green fluorescent protein. Ann. Rev. Biochem. 67, 509–544 (1998)
Andresen, M., Stiel, A.C., Folling, J., Wenzel, D., Schonle, A., Egner, A., Eggeling, C., Hell, S.W., Jakobs, S.: Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat. Biotechnol. 26(9), 1035–1040 (2008)
Andresen, M., Stiel, A.C., Trowitzsch, S., Weber, G., Eggeling, C., Wahl, M.C., Hell, S.W., Jakobs, S.: Structural basis for reversible photoswitching in dronpa. Proc. Natl. Acad. Sci. USA 104(32), 13005–13009 (2007)
Andresen, M., Wahl, M.C., Stiel, A.C., Grater, F., Schafer, L.V., Trowitzsch, S., Weber, G., Eggeling, C., Grubmuller, H., Hell, S.W., Jakobs, S.: Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proc. Natl. Acad. Sci. USA 102(37), 13070–13074 (2005)
Brakemann, T., Weber, G., Andresen, M., Groenhof, G., Stiel, A.C., Trowitzsch, S., Eggeling, C., Grubmuller, H., Hell, S.W., Wahl, M.C., Jakobs, S.: Molecular basis of the light-driven switching of the photochromic fluorescent protein padron. J. Biol. Chem. 285(19), 14603–14609 (2010)
Duan, C., Adam, V., Byrdin, M., Bourgeois, D.: Structural basis of photoswitching in fluorescent proteins. Methods Mol. Biol. 1148, 177–202 (2014)
El Khatib, M., Martins, A., Bourgeois, D., Colletier, J.P., Adam, V.: Rational design of ultrastable and reversibly photoswitchable fluorescent proteins for super-resolution imaging of the bacterial periplasm. Sci. Rep. 6, 18459 (2016)
Hutchison, C.D.M., Cordon-Preciado, V., Morgan, R.M.L., Nakane, T., Ferreira, J., Dorlhiac, G., Sanchez-Gonzalez, A., Johnson, A.S., Fitzpatrick, A., Fare, C., Marangos, J.P., Yoon, C.H., Hunter, M.S., DePonte, D.P., Boutet, S., Owada, S., Tanaka, R., Tono, K., Iwata, S., van Thor, J.J.: X-ray free electron laser determination of crystal structures of dark and light states of a reversibly photoswitching fluorescent protein at room temperature. Int. J. Mol. Sci. 18(9) (2017)
Pletnev, S., Subach, F.V., Dauter, Z., Wlodawer, A., Verkhusha, V.V.: A structural basis for reversible photoswitching of absorbance spectra in red fluorescent protein rsTagRFP. J. Mol. Biol. 417(3), 144–151 (2012)
Bourgeois, D.: Deciphering structural photophysics of fluorescent proteins by kinetic crystallography. Int. J. Mol. Sci. 18(6), 1187 (2017)
Coquelle, N., Sliwa, M., Woodhouse, J., Schiro, G., Adam, V., Aquila, A., Barends, T.R.M., Boutet, S., Byrdin, M., Carbajo, S., De la Mora, E., Doak, R.B., Feliks, M., Fieschi, F., Foucar, L., Guillon, V., Hilpert, M., Hunter, M.S., Jakobs, S., Koglin, J.E., Kovacsova, G., Lane, T.J., Levy, B., Liang, M., Nass, K., Ridard, J., Robinson, J.S., Roome, C.M., Ruckebusch, C., Seaberg, M., Thepaut, M., Cammarata, M., Demachy, I., Field, M., Shoeman, R.L., Bourgeois, D., Colletier, J.P., Schlichting, I., Weik, M.: Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography. Nat. Chem. 10(1), 31–37 (2018)
Warren, M.M., Kaucikas, M., Fitzpatrick, A., Champion, P., Sage, J.T., van Thor, J.J.: Ground-state proton transfer in the photoswitching reactions of the fluorescent protein dronpa. Nat. Commun. 4, 1461 (2013)
Yadav, D., Lacombat, F., Dozova, N., Rappaport, F., Plaza, P., Espagne, A.: Real-time monitoring of chromophore isomerization and deprotonation during the photoactivation of the fluorescent protein dronpa. J. Phys. Chem. B 119(6), 2404–2414 (2015)
Bourgeois, D., Adam, V.: Reversible photoswitching in fluorescent proteins: a mechanistic view. IUBMB Life 64(6), 482–491 (2012)
Petersen, J., Wilmann, P.G., Beddoe, T., Oakley, A.J., Devenish, R.J., Prescott, M., Rossjohn, J.: The 2.0-Å crystal structure of eqFP611, a far red fluorescent protein from the sea anemone entacmaea quadricolor. J. Biol. Chem. 278(45), 44626–44631 (2003)
Brakemann, T., Stiel, A.C., Weber, G., Andresen, M., Testa, I., Grotjohann, T., Leutenegger, M., Plessmann, U., Urlaub, H., Eggeling, C., Wahl, M.C., Hell, S.W., Jakobs, S.: A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat. Biotechnol. 29(10), 942–947 (2011)
Arai, Y., Takauchi, H., Ogami, Y., Fujiwara, S., Nakano, M., Matsuda, T., Nagai, T.: Spontaneously blinking fluorescent protein for simple single laser super-resolution live cell imaging. ACS Chem. Biol. 13(8), 1938–1943 (2018)
Duwe, S., De Zitter, E., Gielen, V., Moeyaert, B., Vandenberg, W., Grotjohann, T., Clays, K., Jakobs, S., Van Meervelt, L., Dedecker, P.: Expression-enhanced fluorescent proteins based on enhanced green fluorescent protein for super-resolution microscopy. ACS Nano 9(10), 9528–9541 (2015)
Colletier, J.P., Sliwa, M., Gallat, F.X., Sugahara, M., Guillon, V., Schiro, G., Coquelle, N., Woodhouse, J., Roux, L., Gotthard, G., Royant, A., Uriarte, L.M., Ruckebusch, C., Joti, Y., Byrdin, M., Mizohata, E., Nango, E., Tanaka, T., Tono, K., Yabashi, M., Adam, V., Cammarata, M., Schlichting, I., Bourgeois, D., Weik, M.: Serial femtosecond crystallography and ultrafast absorption spectroscopy of the photoswitchable fluorescent protein IrisFP. J. Phys. Chem. Lett. 7(5), 882–887 (2016)
Stiel, A.C., Andresen, M., Bock, H., Hilbert, M., Schilde, J., Schonle, A., Eggeling, C., Egner, A., Hell, S.W., Jakobs, S.: Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys. J. 95(6), 2989–2997 (2008)
Lukyanov, K.A., Fradkov, A.F., Gurskaya, N.G., Matz, M.V., Labas, Y.A., Savitsky, A.P., Markelov, M.L., Zaraisky, A.G., Zhao, X., Fang, Y., Tan, W., Lukyanov, S.A.: Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J. Biol. Chem. 275(34), 25879–25882 (2000)
Tiwari, D.K., Arai, Y., Yamanaka, M., Matsuda, T., Agetsuma, M., Nakano, M., Fujita, K., Nagai, T.: A fast- and positively photoswitchable fluorescent protein for ultralow-laser-power resolft nanoscopy. Nat. Methods 12(6), 515–518 (2015)
Lacombat, F., Plaza, P., Plamont, M.A., Espagne, A.: Photoinduced chromophore hydration in the fluorescent protein dreiklang is triggered by ultrafast excited-state proton transfer coupled to a low-frequency vibration. J. Phys. Chem. Lett. 8(7), 1489–1495 (2017)
Jensen, N.A., Danzl, J.G., Willig, K.I., Lavoie-Cardinal, F., Brakemann, T., Hell, S.W., Jakobs, S.: Coordinate-targeted and coordinate-stochastic super-resolution microscopy with the reversibly switchable fluorescent protein dreiklang. Chemphyschem 15(4), 756–762 (2014)
Cranfill, P.J., Sell, B.R., Baird, M.A., Allen, J.R., Lavagnino, Z., de Gruiter, H.M., Kremers, G.J., Davidson, M.W., Ustione, A., Piston, D.W.: Quantitative assessment of fluorescent proteins. Nat. Methods 13(7), 557–562 (2016)
Heppert, J.K., Dickinson, D.J., Pani, A.M., Higgins, C.D., Steward, A., Ahringer, J., Kuhn, J.R., Goldstein, B.: Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol. Biol. Cell 27(22), 3385–3394 (2016)
Pennacchietti, F., Serebrovskaya, E.O., Faro, A.R., Shemyakina II, Bozhanova, N.G., Kotlobay, A.A., Gurskaya, N.G., Boden, A., Dreier, J., Chudakov, D.M., Lukyanov, K.A., Verkhusha, V.V., Mishin, A.S., Testa, I.: Fast reversibly photoswitching red fluorescent proteins for live-cell resolft nanoscopy. Nat. Methods 15(8), 601–604 (2018)
Wang, S., Chen, X., Chang, L., Ding, M., Xue, R., Duan, H., Sun, Y.: Gmars-t enabling multimodal subdiffraction structural and functional fluorescence imaging in live cells. Anal. Chem. 90(11), 6626–6634 (2018)
Faro, A.R., Carpentier, P., Jonasson, G., Pompidor, G., Arcizet, D., Demachy, I., Bourgeois, D.: Low-temperature chromophore isomerization reveals the photoswitching mechanism of the fluorescent protein padron. J. Am. Chem. Soc. 133(41), 16362–16365 (2011)
Habuchi, S., Ando, R., Dedecker, P., Verheijen, W., Mizuno, H., Miyawaki, A., Hofkens, J.: Reversible single-molecule photoswitching in the gfp-like fluorescent protein dronpa. Proc. Natl. Acad. Sci. USA 102(27), 9511–9516 (2005)
Chmyrov, A., Leutenegger, M., Grotjohann, T., Schonle, A., Keller-Findeisen, J., Kastrup, L., Jakobs, S., Donnert, G., Sahl, S.J., Hell, S.W.: Achromatic light patterning and improved image reconstruction for parallelized resolft nanoscopy. Sci. Rep. 7, 44619 (2017)
Zhang, X., Zhang, M., Li, D., He, W., Peng, J., Betzig, E., Xu, P.: Highly photostable, reversibly photoswitchable fluorescent protein with high contrast ratio for live-cell superresolution microscopy. Proc. Natl. Acad. Sci. USA 113(37), 10364–10369 (2016)
Hell, S.W.: Microscopy and its focal switch. Nat. Methods 6(1), 24–32 (2009)
Duan, C., Adam, V., Byrdin, M., Ridard, J., Kieffer-Jaquinod, S., Morlot, C., Arcizet, D., Demachy, I., Bourgeois, D.: Structural evidence for a two-regime photobleaching mechanism in a reversibly switchable fluorescent protein. J. Am. Chem. Soc. 135(42), 15841–15850 (2013)
Grotjohann, T., Testa, I., Leutenegger, M., Bock, H., Urban, N.T., Lavoie-Cardinal, F., Willig, K.I., Eggeling, C., Jakobs, S., Hell, S.W.: Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature 478(7368), 204–208 (2011)
Ando, R., Flors, C., Mizuno, H., Hofkens, J., Miyawaki, A.: Highlighted generation of fluorescence signals using simultaneous two-color irradiation on dronpa mutants. Biophys. J. 92(12), L97–L99 (2007)
Stiel, A.C., Trowitzsch, S., Weber, G., Andresen, M., Eggeling, C., Hell, S.W., Jakobs, S., Wahl, M.C.: 1.8 a bright-state structure of the reversibly switchable fluorescent protein dronpa guides the generation of fast switching variants. Biochem. J. 402(1), 35–42 (2007)
Zhou, X.X., Lin, M.Z.: Photoswitchable fluorescent proteins: ten years of colorful chemistry and exciting applications. Curr. Opin. Chem. Biol. 17(4), 682–690 (2013)
Ando, R., Mizuno, H., Miyawaki, A.: Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306(5700), 1370–1373 (2004)
Subach, F.V., Zhang, L., Gadella, T.W., Gurskaya, N.G., Lukyanov, K.A., Verkhusha, V.V.: Red fluorescent protein with reversibly photoswitchable absorbance for photochromic fret. Chem. Biol. 17(7), 745–755 (2010)
Zhou, X.X., Chung, H.K., Lam, A.J., Lin, M.Z.: Optical control of protein activity by fluorescent protein domains. Science 338(6108), 810–814 (2012)
Vetschera, P., Mishra, K., Fuenzalida-Werner, J.P., Chmyrov, A., Ntziachristos, V., Stiel, A.C.: Characterization of reversibly switchable fluorescent proteins in optoacoustic imaging. Anal. Chem. 90(17), 10527–10535 (2018)
Geissbuehler, S., Sharipov, A., Godinat, A., Bocchio, N.L., Sandoz, P.A., Huss, A., Jensen, N.A., Jakobs, S., Enderlein, J., Gisou van der Goot, F., Dubikovskaya, E.A., Lasser, T., Leutenegger, M.: Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nat. Commun. 5, 5830 (2014)
Zhang, X., Chen, X., Zeng, Z., Zhang, M., Sun, Y., Xi, P., Peng, J., Xu, P.: Development of a reversibly switchable fluorescent protein for super-resolution optical fluctuation imaging (SOFI). ACS Nano 9(3), 2659–2667 (2015)
Danzl, J.G., Sidenstein, S.C., Gregor, C., Urban, N.T., Ilgen, P., Jakobs, S., Hell, S.W.: Coordinate-targeted fluorescence nanoscopy with multiple off states. Nat. Photonics 10, 122 (2016)
Willig, K.I., Stiel, A.C., Brakemann, T., Jakobs, S., Hell, S.W.: Dual-label sted nanoscopy of living cells using photochromism. Nano Lett. 11(9), 3970–3973 (2011)
Wang, S., Chen, X., Chang, L., Xue, R., Duan, H., Sun, Y.: Gmars-Q enables long-term live-cell parallelized reversible saturable optical fluorescence transitions nanoscopy. ACS Nano 10(10), 9136–9144 (2016)
Adam, V., Moeyaert, B., David, C.C., Mizuno, H., Lelimousin, M., Dedecker, P., Ando, R., Miyawaki, A., Michiels, J., Engelborghs, Y., Hofkens, J.: Rational design of photoconvertible and biphotochromic fluorescent proteins for advanced microscopy applications. Chem. Biol. 18(10), 1241–1251 (2011)
Chmyrov, A., Keller, J., Grotjohann, T., Ratz, M., d’Este, E., Jakobs, S., Eggeling, C., Hell, S.W.: Nanoscopy with more than 100,000 ‘doughnuts’. Nat. Methods 10(8), 737–740 (2013)
Chang, H., Zhang, M., Ji, W., Chen, J., Zhang, Y., Liu, B., Lu, J., Zhang, J., Xu, P., Xu, T.: A unique series of reversibly switchable fluorescent proteins with beneficial properties for various applications. Proc. Natl. Acad. Sci. USA 109(12), 4455–4460 (2012)
Fuchs, J., Bohme, S., Oswald, F., Hedde, P.N., Krause, M., Wiedenmann, J., Nienhaus, G.U.: A photoactivatable marker protein for pulse-chase imaging with superresolution. Nat. Methods 7(8), 627–630 (2010)
Lavoie-Cardinal, F., Jensen, N.A., Westphal, V., Stiel, A.C., Chmyrov, A., Bierwagen, J., Testa, I., Jakobs, S., Hell, S.W.: Two-color resolft nanoscopy with green and red fluorescent photochromic proteins. Chemphyschem 15(4), 655–663 (2014)
Patterson, G.H., Knobel, S.M., Sharif, W.D., Kain, S.R., Piston, D.W.: Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73(5), 2782–2790 (1997)
Testa, I., Urban, N.T., Jakobs, S., Eggeling, C., Willig, K.I., Hell, S.W.: Nanoscopy of living brain slices with low light levels. Neuron 75(6), 992–1000 (2012)
Gurskaya, N.G., Verkhusha, V.V., Shcheglov, A.S., Staroverov, D.B., Chepurnykh, T.V., Fradkov, A.F., Lukyanov, S., Lukyanov, K.A.: Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 24(4), 461–465 (2006)
Wiedenmann, J., Ivanchenko, S., Oswald, F., Schmitt, F., Rocker, C., Salih, A., Spindler, K.D., Nienhaus, G.U.: EosFP, a fluorescent marker protein with uv-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 101(45), 15905–15910 (2004)
Wiedenmann, J., Gayda, S., Adam, V., Oswald, F., Nienhaus, K., Bourgeois, D., Nienhaus, G.U.: From EosFP to mIrisFP: structure-based development of advanced photoactivatable marker proteins of the GFP-family. J. Biophotonics 4(6), 377–390 (2011)
Moeyaert, B., Bich, N.N., De Zitter, E., Rocha, S., Clays, K., Mizuno, H., van Meervelt, L., Hofkens, J., Dedecker, P.: Green-to-red photoconvertible dronpa mutant for multimodal super-resolution fluorescence microscopy. ACS Nano 8(2), 1664–1673 (2014)
McKinney, S.A., Murphy, C.S., Hazelwood, K.L., Davidson, M.W., Looger, L.L.: A bright and photostable photoconvertible fluorescent protein. Nat. Methods 6(2), 131–133 (2009)
Zhang, M., Chang, H., Zhang, Y., Yu, J., Wu, L., Ji, W., Chen, J., Liu, B., Lu, J., Liu, Y., Zhang, J., Xu, P., Xu, T.: Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9(7), 727–729 (2012)
Wang, S., Moffitt, J.R., Dempsey, G.T., Xie, X.S., Zhuang, X.: Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA 111(23), 8452–8457 (2014)
Wang, S., Shuai, Y., Sun, C., Xue, B., Hou, Y., Su, X., Sun, Y.: Lighting up live cells with smart genetically encoded fluorescence probes from gmars family. ACS Sens. 3(11), 2269–2277 (2018)
Griesbeck, O., Baird, G.S., Campbell, R.E., Zacharias, D.A., Tsien, R.Y.: Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276(31), 29188–29194 (2001)
Hofmann, M., Eggeling, C., Jakobs, S., Hell, S.W.: Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci. USA 102(49), 17565–17569 (2005)
Merzlyak, E.M., Goedhart, J., Shcherbo, D., Bulina, M.E., Shcheglov, A.S., Fradkov, A.F., Gaintzeva, A., Lukyanov, K.A., Lukyanov, S., Gadella, T.W., Chudakov, D.M.: Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 4(7), 555–557 (2007)
Ilgen, P., Grotjohann, T., Jans, D.C., Kilisch, M., Hell, S.W., Jakobs, S.: Resolft nanoscopy of fixed cells using a z-domain based fusion protein for labelling. PLoS One 10(9), e0136233 (2015)
Masullo, L.A., Boden, A., Pennacchietti, F., Coceano, G., Ratz, M., Testa, I.: Enhanced photon collection enables four dimensional fluorescence nanoscopy of living systems. Nat. Commun. 9(1), 3281 (2018)
Wang, S., Ding, M., Chen, X., Chang, L., Sun, Y.: Development of bimolecular fluorescence complementation using rsEGFP2 for detection and super-resolution imaging of protein-protein interactions in live cells. Biomed. Opt. Express 8(6), 3119–3131 (2017)
Ratz, M., Testa, I., Hell, S.W., Jakobs, S.: Crispr/cas9-mediated endogenous protein tagging for resolft super-resolution microscopy of living human cells. Sci. Rep. 5, 9592 (2015)
Bohm, U., Hell, S.W., Schmidt, R.: 4Pi-resolft nanoscopy. Nat. Commun. 7, 10504 (2016)
Testa, I., D’Este, E., Urban, N.T., Balzarotti, F., Hell, S.W.: Dual channel resolft nanoscopy by using fluorescent state kinetics. Nano Lett. 15(1), 103–106 (2015)
Li, D., Shao, L., Chen, B.C., Zhang, X., Zhang, M., Moses, B., Milkie, D.E., Beach, J.R., Hammer, J.A. 3rd, Pasham, M., Kirchhausen, T., Baird, M.A., Davidson, M.W., Xu, P., Betzig, E.: Advanced imaging. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349(6251), aab3500 (2015)
Hoyer, P., de Medeiros, G., Balazs, B., Norlin, N., Besir, C., Hanne, J., Krausslich, H.G., Engelhardt, J., Sahl, S.J., Hell, S.W., Hufnagel, L.: Breaking the diffraction limit of light-sheet fluorescence microscopy by resolft. Proc. Natl. Acad. Sci. USA 113(13), 3442–3446 (2016)
Gregor, C., Sidenstein, S.C., Andresen, M., Sahl, S.J., Danzl, J.G., Hell, S.W.: Novel reversibly switchable fluorescent proteins for RESOLFT and STED nanoscopy engineered from the bacterial photoreceptor YtvA. Sci. Rep. 8(1), 2724 (2018)
Roubinet, B., Bossi, M.L., Alt, P., Leutenegger, M., Shojaei, H., Schnorrenberg, S., Nizamov, S., Irie, M., Belov, V.N., Hell, S.W.: Carboxylated photoswitchable diarylethenes for biolabeling and super-resolution resolft microscopy. Angew. Chem. Int. Ed. Engl. 55(49), 15429–15433 (2016)
Kwon, J., Hwang, J., Park, J., Han, G.R., Han, K.Y., Kim, S.K.: Resolft nanoscopy with photoswitchable organic fluorophores. Sci. Rep. 5, 17804 (2015)
Lychagov, V.V., Shemetov, A.A., Jimenez, R., Verkhusha, V.V.: Microfluidic system for in-flow reversible photoswitching of near-infrared fluorescent proteins. Anal. Chem. 88(23), 11821–11829 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Jensen, N.A., Jansen, I., Kamper, M., Jakobs, S. (2020). Reversibly Switchable Fluorescent Proteins for RESOLFT Nanoscopy. In: Salditt, T., Egner, A., Luke, D.R. (eds) Nanoscale Photonic Imaging. Topics in Applied Physics, vol 134. Springer, Cham. https://doi.org/10.1007/978-3-030-34413-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-34413-9_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34412-2
Online ISBN: 978-3-030-34413-9
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)