Abstract
The quality of sleep has a significant impact on health and life. This study adopts the structure of hierarchical classification to develop an automatic sleep stage classification system using ballistocardiogram (BCG) signals. A leave-one-subject-out cross validation (LOSO-CS) procedure is used for testing classification performance. Convolutional Neural Networks (CNNs), Long Short-Term Memory (LSTM), and Deep Neural Networks DNNs are complementary in their modeling capabilities; while CNNs have the advantage of reducing frequency variations, LSTMs are good at temporal modeling. A transfer learning (TL) technique is used to pre-train our CNN model on posture data and then fine-tune it on the sleep stage data. We used a ballistocardiography (BCG) bed sensor to collect both posture and sleep stage data to provide a non-invasive, in-home monitoring system that tracks changes in health of the subjects over time. Polysomnography (PSG) data from a sleep lab was used as the ground truth for sleep stages, with the emphasis on three sleep stages, specifically, awake, rapid eye movement (REM) and non-REM sleep (NREM). Our results show an accuracy of 95.3%, 84% and 93.1% for awake, REM and NREM respectively on a group of patients from the sleep lab.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Sleep is a critical physiological phenomenon for recovery from mental and physical fatigue. Lately, there has been much interest in the quality of sleep, and research is actively underway. In particular, it is vital to have a repetitive and regular sleep cycle for good sleep. Nevertheless, it takes much time to determine sleep stages using physiological signals by experts. A person with a sleep disorder such as apnea will stop breathing for a while throughout sleep. If it regularly happens, sleep disorders can be risky for health. An early step in diagnosing these disorders is the classification of sleep stages [4, 13].
Unfortunately, sleep disorders have been affecting many people around the world in different ways. Whatever the cause of these disorders, the consequences can be severe. The quality of sleep depends on the number and order of these stages. The names of these stages, are Wake, Non-REM1, Non-REM2, Non-REM3, and REM. Detection of any sleep disorder, such as sleep apnea, insomnia, or narcolepsy, requires a correct staging of sleep [2, 18].
Classification of sleep stages is also essential for managing the quality of sleep. Sleep studies depend on manual scoring of sleep stages from raw polysomnography signals, which is a tedious visual task. Thus, research efforts to develop an automatic sleep stage scoring based on machine learning techniques have been carried out in the last several years [19]. Convolutional neural networks (CNN) [14] and Long-Short Term Memory Recurrent Neural Networks (LSTM) [15] provide an interesting framework for automated classification of sleep based on raw waveforms. In the past few years, Deep Neural Networks (DNNs) have accomplished tremendous success for time series tasks compared to traditional machine learning systems. Recently, further improvements over DNNs have been obtained with alternative types of neural network architectures. CNNs, LSTMs, and DNNs are individually limited in their modeling capabilities, and we believe that time series data classification can be improved by combining these networks in a unified framework. The classification of time series signals presents many challenges that make it a uniquely difficult problem in machine learning. Many feature extraction approaches in time series face issues related to the non-stationary nature of the signal when the probability distribution does not change over time. Accordingly, features such as mean and variance will not change. Furthermore, the physiological signals are very noisy, being susceptible to factors such as posture, mood, physical movement, and external noise [8]. Lack of comparability between experiments is another issue that can be faced in this field. Unlike in image classification, there are no standard time series datasets used as performance benchmarks [16, 20]. Some approaches use models for individuals, while others try to make a general model, training, and testing with samples from all individuals at one time.
In this paper, we propose a method for classifying sleep stages based on the CNN, LSTM and DNN with the help of transfer learning. More specifically, we use a transfer learning technique to train our network model with sleep posture data for 56 subjects (source dataset) and use it for sleep stage classification (target data). The sleep data was obtained from 5 subjects and it was collected in the Boone Hospital Center (BHC) in Columbia, MO, USA under the University of Missouri IRB approval, project number 2008526. The main contribution of this work is developing a new deep model architecture that utilizes CNNs and LSTMs to classify sleep stage data. The CNNs are trained to learn filters that extract time-invariant features from the BCG signals while the LSTMs are trained to encode temporal information such as sleep stage transition rules.
2 Sensors and Datasets
A home monitoring system using a ballistocardiography (BCG) hydraulic sensor has been developed to monitor sleep at home (see Fig. 1). The hydraulic bed sensor has been developed at the Center for Eldercare and Rehabilitation Technology (CERT) at the University of Missouri. A BCG device provides a noninvasive, low-cost, and robust solution for capturing physiological parameters such as heart rate and respiration rate, during sleep [6, 10, 11]. The BCG sensor has four transducers. Each transducer is composed of a water tube with a pressure sensor placed at one end. The water tube is 50 cm long and 6 cm wide and it is filled with about 0.4 liters of water. The BCG sensor is placed under the mattress, parallel to the body direction, to provide sleeping comfort and not to disturb a person’s normal sleep pattern.
The pressure outputs are coupled to a Maxim MAX7401 which is a filtering circuit that consists of a 741 operational amplifier and an 8th-order integrated Bessel filter [3, 9]. The four-channel signal is sampled and quantized to 12-bit precision. The BCG signal acquired from the sensor is superimposed over the respiration signal. The four matching transducers are independent; consequently, the data quality collected by those transducers might vary depending on the subject’s sleeping position, type of bed (e.g., material, thickness, etc.) and the physical characteristics of the subject such as age and body mass index (BMI). In this study, two kinds of datasets have been collected utilizing our bed sensor: sleep posture and sleep stages.
2.1 Posture Dataset
A total of 56 young healthy subjects (About 75.8% of males and 24.2% of females) were asked to lie still in our lab for one minute on each of the four main postures, supine, prone, left lateral and right lateral. The data collection procedure was approved by the University of Missouri Institutional Review Board (MUIRB). The bed sensor produced data sampled at 100 Hz. The subjects age ranged between 18 and 49 years (mean 29.27 years); the weight ranged between 48 and 184 kg (mean 77.40 kg); the height ranged between 100 and 190 cm (mean 173.04 cm); the average body mass index was 27.34 kg/m\(^{2}\), ranged between 18.1 and 184 kg/m\(^{2}\).
2.2 Sleep Stage Dataset
Sleep stage data was collected from consenting patients by a sleep-credentialed physician during the regularly scheduled PSG studies conducted in the sleep lab of the Boone Hospital Center (BHC) in Columbia, MO, USA. In addition to the regular PSG equipment, we placed our hydraulic bed sensor under the study bed mattress. The scoring system for staging the sleep was based on the American Academy of Sleep Medicine Manual (AASM) [1], which is the standard for scoring sleep stages and it provides guidelines for associated events during sleep. Sleep stage scoring in 30-second epochs is required for the AASM protocol. A program produced by Natus SleepWorks (Natus Medical Inc., San Carlos, CA, USA) was used in the BHC sleep lab to assist the staff in monitoring a patient throughout the night. SleepWorks not only collects the PSG data but also performs a video recording of the entire night. The technician can view the patient’s sleep video if they have any uncertainty about the data. SleepWorks can also provide an initial analysis of the collected data and can generate a report, which helps the sleep physicians who make treatment recommendations.
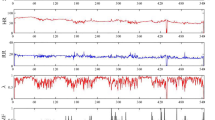
The BHC sleep lab provides de-identified polysomnography (PSG) data with a sample rate of 256 Hz. PSG is a multiparametric recording method used in sleep labs to monitor physiological changes throughout sleep. It is a consistent tool for diagnosing sleep disorders, and it can similarly help adjust treatment. Moreover, a technician scores each patient’s clinical events (e.g., limb movements, respiratory and cardiac events, and arousals) using the American Academy of Sleep Medicine (AASM) standards. PSG is used in this research as ground truth for our bed sensor signals. The hypnogram is one PSG outcome that displays the sleep stages as a function of time (Fig. 2). Sleep stages are annotated in 30-second epochs. From top to bottom, the sleep stages are Wake (W), REM (R), NREM1 (N1), NREM2 (N2), and NREM3 (N3). For the patient shown in Fig. 2, 742 epochs were monitored during sleep. REM sleep happened between epoch 392 and epoch 485, also highlighted in red.
Those who participated in the sleep study are likely to have a sleep disorder. The partial and the complete collapse of the airways are called hypopnea and apnea, respectively. Lack of airflow will affect breathing patterns and then influence sleep stage sequence. The Apnea-hypopnea index (AHI) represents the number of apnea and hypopnea events per hour during sleep. Based on the AASM, the mild sleep apnea AHI is between 5 and 15. In this study, we did not use the data collected from patients with severe apnea symptoms. For this study, we selected five sleep lab patients with a low Apnea-hypopnea index (AHI), so that each one has sufficient REM, NREM, and Wake sleep stages during the night.
2.3 Data Preprocessing
We present the preprocessing steps for our data. For the posture data, we truncate the signals utilizing the percentile method [21] to remove any noise that was introduced due to the desynchronization between the time of leaving the bed and stopping recording the signal. After denoising, we normalize the data by utilizing standard score normalization technique. Then we down-sample the data to 100 Hz and shuffle it to prepare it for net training. To account for the varying length of the signals, we divide the preprocessed bed sensor signals into 5-second segments with 80% overlap. Each output label corresponds to a segment of the input. Together the output labels cover the full sequence. The above two algorithms are applied on posture and sleep stage data. Another algorithm is applied for sleep stages only, which are the rules from [22] that help to improve the accuracy about 1%. These rules can be illustrated as followed:
-
For three consecutive 30s epochs, if the center one is not the same as the other two and the other two are the same, change the center one to the same stage as the other two epochs. However, the center epoch is not changed if it is an Awake stage.
-
For every three 30s epochs, if the sleep stages are all different, the center epoch is removed from the recording. Similarly, this rule does not apply to the Awake stage.
-
If REM stages show up in the first hour, these stages are removed from the recording.
3 Architecture Design
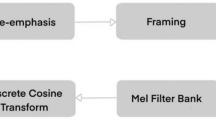
We propose a new architecture to classify BCG bed sensor data (see Fig. 3.). The network takes as input the windows of time series of filtered BCG signal and outputs a sequence of label predictions. The basic block is a convolution layer followed by a batch normalization layer [7], Rectified Linear Unit (ReLU) activation layer, and max-pooling layer. We similarly employ shortcut connections to those found in the Residual Network architecture [5] to make the optimization of such a network manageable. The shortcut connections between neural network layers enhance training by permitting by permitting information to propagate well in deep neural networks. The convolutional layers in the first and second blocks have 8 filters; this number of filters doubles in successive blocks until it reaches 32 in the last block. Moreover, the Max Pooling layer helps to subsample the input to become one-fourth of the input sample at the top layer since Max Pool is set to size two. When a block subsamples the input, the corresponding shortcut connections also subsample their input using a Max Pooling operation with the same subsample size. We next pass the CNN block output to LSTM layers with 128 units, which are appropriate for modeling the signal in time.
Regularization techniques are employed in our architecture to reduce overfitting effects during training. Regularization techniques help keep the model from becoming too complex and specific to the training data, thus reducing the tendency to overfit. In this paper, two regularization techniques were used. The first technique was a dropout, which randomly sets the input value to zero with a certain probability [17]. A probability of 0.5 was used in the dropout layer after LSTM, as shown in Fig. 3. The second technique was L2 weight decay, which adds a penalty term into the loss function. L2 regularization is a typical technique used in many optimization methods, in which the squared sum of the weights is applied as a penalty to the optimization function. In essence, this weighs the advantage of increased classification of the training data against model complexity. By preventing the model from becoming too complicated, memorization of the training data is reduced, and a more generalizable model is developed.
4 Training and Experimental Results
Transfer learning is employed in the experiments of this paper to classify sleep stages. Transfer learning is a technique wherein the knowledge gained from training a model on a dataset (source) can be reused as a starting point, when fine-tuning the model on another dataset (target). This is usually done when the target dataset is not as rich as the source, and hence, a complex deep network would have more trouble training only on the target. Consequently, we split the network model training into two distinct phases. First, our model is trained on the class-balanced training posture dataset of 56 people using five-fold cross-validation. Then, the network weights obtained in the first phase are used as an initialization for the second phase of training on sleep stage data subjects.
We conducted LOSO-CS to fine-tune the sleep stage data. Each time (fold), one patient was left out for testing while the remaining four patients were used to train the DNN networks. In the testing part, we utilize a hierarchical classification scheme for our automatic sleep stage classification system. This structure is composed of two layers. The first layer separates wake from the sleep, which is a union of REM and NREM, in a binary classification problem. Next, all the epochs classified as sleep are fed into the next layer to classify the REM and NREM epochs further. The last block of the architecture, shown in Fig. 3 is used to fine-tune the sleep stage data. In the first phase, the optimal hyperparameters for the model are carefully chosen for a given dataset. The model is at that time fine-tuned on the given dataset with these hyperparameter settings.
Training overnight raw sleep stage data using DNN is time-consuming. Moreover, the ground truth of the posture data is more reliable than the ground truth of sleep stage data. Deep learning models with transfer learning can, however, successfully converge on short-time data (sleep posture data) split across multiple subjects. By training a model over multiple posture data subjects, we obtained an initialization that is then used for fine-tuning our model on the sleep stage data. Based on Williams et al. [20], this process is hypothesized to make convergence on an individual’s data more likely; it should also provide stronger general filters in the first layers of the neural network.
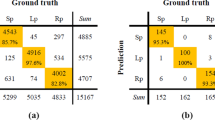
Figure 4(a) and (b) show the average of fine-tuned training five-fold LOSO-CS sleep stage data to classify wake from sleep and to classify REM from NREM respectively. Figure 4(c) shows the average of hierarchical fine-tuned testing five-fold LOSO-CS sleep stage data. Clearly, our model was able to classify the sleep stage classes better than the results reported by Yi, et al. [23] which were 79.9%, 78.8%, and 88.8% sensitivity for wake, REM, and NREM respectively; the previous work has been done using the same data and the same sensor, but with traditional machine learning algorithms. Table 1 shows the percentage of epochs that are correctly classified for each class Wake, REM, and NREM comparing to the percentage ground truth. The sleep stage agreement was 3.64%, 2.76%, and 8.920% for the Wake, REM, and NREM, respectively. This agreement result is much better than the agreement reported by Rosenberg et al. [12], which was 82.6% as the highest agreement averaged for REM and 63.0% as the lowest agreement averaged for NREM. From the results shown in Fig. 4, and Table 4, it is worth to mention the misclassification of the Wake as Sleep in the first layer in the hierarchical method is fed into the second layer, which has a cumulative impact on the second layer classification.
5 Conclusion
In this paper, we have developed a deep learning-based hierarchical classification method for automatic sleep stage classification based on BCG data. The deep learning model is a state-of-the-art algorithm that consists of a stacked CNN-LSTM model. The proposed model was trained using a transfer learning approach that achieved a significantly improved performance in comparison to similar studies. The results of the leave-one-out cross-validation strategy showed potential in automatically classifying sleep stage epochs. Our sleep monitoring system based on the BCG can indeed provide a more natural way of diagnosing sleep problems, and make long-term sleep monitoring possible. However, to validate our algorithms we used a selected, balanced dataset with a reduced level of noise. We are currently working to increase the robustness of our methodology and validate it for patients with reduced REM and high AHI.
References
Berry, R.B., Brooks, R., Gamaldo, C.E., Harding, S.M., Marcus, C.L., Vaughn, B.V., et al.: The AASM manual for the scoring of sleep and associated events. Rules, Terminology and Technical Specifications. American Academy of Sleep Medicine, Darien, Illinois, vol. 176 (2012)
Dursun, M., Gunes, S., Ozsen, S., Yosunkaya, S.: Comparison of artificial immune clustering with fuzzy c-means clustering in the sleep stage classification problem. In: 2012 International Symposium on Innovations in Intelligent Systems and Applications, pp. 1–4. IEEE (2012)
Gargees, R., Keller, J., Popescu, M.: Early illness recognition in older adults using transfer learning. In: 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1012–1016. IEEE (2017)
Giri, E.P., Arymurthy, A.M., Fanany, M.I., Wijaya, S.K.: Sleep stages classification using shallow classifiers. In: 2015 International Conference on Advanced Computer Science and Information Systems (ICACSIS), pp. 297–301. IEEE (2015)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Heise, D., Rosales, L., Sheahen, M., Su, B.Y., Skubic, M.: Non-invasive measurement of heartbeat with a hydraulic bed sensor progress, challenges, and opportunities. In: 2013 IEEE International Instrumentation and Measurement Technology Conference (I2MTC), pp. 397–402. IEEE (2013)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on Machine Learning, vol. 37, pp. 448–456 (2015)
Kevric, J., Subasi, A.: Comparison of signal decomposition methods in classification of EEG signals for motor-imagery BCI system. Biomed. Signal Process. Control 31, 398–406 (2017)
Lydon, K., et al.: Robust heartbeat detection from in-home ballistocardiogram signals of older adults using a bed sensor. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 7175–7179. IEEE (2015)
Rosales, L., Skubic, M., Heise, D., Devaney, M.J., Schaumburg, M.: Heartbeat detection from a hydraulic bed sensor using a clustering approach. In: 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 2383–2387. IEEE (2012)
Rosales, L., Su, B.Y., Skubic, M., Ho, K.: Heart rate monitoring using hydraulic bed sensor ballistocardiogram 1. J. Ambient. Intell. Smart Environ. 9(2), 193–207 (2017)
Rosenberg, R.S., Van Hout, S.: The american academy of sleep medicine inter-scorer reliability program: sleep stage scoring. J. Clin. Sleep Med. 9(01), 81–87 (2013)
Sadek, I., Biswas, J., Abdulrazak, B.: Ballistocardiogram signal processing: a review. Health Inf. Sci. Syst. 7(1), 10 (2019)
Sainath, T.N., Vinyals, O., Senior, A., Sak, H.: Convolutional, long short-term memory, fully connected deep neural networks. In: 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 4580–4584. IEEE (2015)
Sak, H., Senior, A., Beaufays, F.: Long short-term memory recurrent neural network architectures for large scale acoustic modeling. In: Fifteenth Annual Conference of the International Speech Communication Association (2014)
Schmidt, S., Eich, G., Hanquinet, S., Tschäppeler, H., Waibel, P., Gudinchet, F.: Extra-osseous involvement of langerhans’ cell histiocytosis in children. Pediatr. Radiol. 34(4), 313–321 (2004)
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
Tuominen, J., Peltola, K., Saaresranta, T., Valli, K.: Sleep parameter assessment accuracy of a consumer home sleep monitoring ballistocardiograph beddit sleep tracker: a validation study. J. Clin. Sleep Med. 15(03), 483–487 (2019)
Vilamala, A., Madsen, K.H., Hansen, L.K.: Deep convolutional neural networks for interpretable analysis of eeg sleep stage scoring. In: 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP), pp. 1–6. IEEE (2017)
Williams, J.M.: Deep learning and transfer learning in the classification of EEG signals (2017)
Wu, M.H., Chang, E.J., Chu, T.H.: Personalizing a generic ECG heartbeat classification for arrhythmia detection: a deep learning approach. In: 2018 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR). IEEE (2018)
Yang, J., Keller, J.M., Popescu, M., Skubic, M.: Sleep stage recognition using respiration signal. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 2843–2846. IEEE (2016)
Yi, R., Enayati, M., Keller, J., Popescu, M., Skubic, M.: Non-invasive in-home sleep stage classification using a ballistocardiography bed sensor. In: 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE (2019)
Acknowledgments
We would like to show our gratitude to all staff of Boone Hospital Center Sleep Center at Columbia, Missouri, with a special thanks to Mr. Custer for his patient and helpful advice.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this paper
Cite this paper
Gargees, R., Keller, J.M., Popescu, M., Skubic, M. (2019). Non-invasive Classification of Sleep Stages with a Hydraulic Bed Sensor Using Deep Learning. In: Pagán, J., Mokhtari, M., Aloulou, H., Abdulrazak, B., Cabrera, M. (eds) How AI Impacts Urban Living and Public Health. ICOST 2019. Lecture Notes in Computer Science(), vol 11862. Springer, Cham. https://doi.org/10.1007/978-3-030-32785-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-32785-9_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-32784-2
Online ISBN: 978-3-030-32785-9
eBook Packages: Computer ScienceComputer Science (R0)