Abstract
This chapter contributes towards advancing finger vein template protection research by presenting the first analysis on the suitability of the BioHashing template protection scheme for finger vein verification systems, in terms of the effect on the system’s recognition performance. Our results show the best performance when BioHashing is applied to finger vein patterns extracted using the Wide Line Detector (WLD) and Repeated Line Tracking (RLT) feature extractors, and the worst performance when the Maximum Curvature (MC) extractor is used. The low recognition performance in the Stolen Token scenario is shown to be improvable by increasing the BioHash length; however, we demonstrate that the BioHash length is constrained in practice by the amount of memory required for the projection matrix. So, WLD finger vein patterns are found to be the most promising for BioHashing purposes due to their relatively small feature vector size, which allows us to generate larger BioHashes than is possible for RLT or MC feature vectors. In addition, we also provide an open-source implementation of a BioHash-protected finger vein verification system based on the WLD, RLT and MC extractors, so that other researchers can verify our findings and build upon our work.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- BioHashing
- Finger veins
- Biometric template protection
- Wide Line Detector
- Repeated Line Tracking
- Maximum Curvature
- EU General Data Protection Regulation (GDPR)
- UTFVP
1 Introduction
As our world is transforming into an interconnected network of individuals and devices, we are beginning to realise that current data protection mechanisms are becoming inadequate to meet our growing security needs. Traditional security mechanisms, such as passwords and access cards, are no longer sufficient for establishing an individual’s true identity, which is why we are turning to biometrics for stronger identity assurance. While the unique link between an individual and their biometric characteristics is the very fact that makes biometric authentication so reliable, it is this same aspect of biometrics that makes this authentication factor vulnerable. For this reason, the past decade has seen the emergence of a new field of research into developing effective biometric template protection strategies to secure biometric features during storage and transmission in an authentication system.Footnote 1 Research in this area is particularly important in light of the recent EU General Data Protection Regulation (GDPR),Footnote 2 which legally obliges users of biometric data to exercise caution in processing and storing this data to protect individuals’ digital identities.
A recent review paper on biometric template protection by Sandhya and Prasad [1] shows that, between the years 2005 to 2016, the smallest amount of effort has been invested into developing protection mechanisms for finger veins. Nevertheless, finger vein recognition has increased in popularity over the past few years, with several companies having already deployed finger vein recognition systems for public use, e.g. M2SYS, Idemia, Hitachi and NEC. This suggests that there is an urgent need to direct our attention towards researching effective mechanisms for protecting finger vein templates.
Although the finger vein template protection field is still in its infancy, a number of methods have been proposed in the literature. For example, in one of the earliest approaches towards finger vein template protection [2], the finger vein pattern image is first transformed using the Number Theoretic Transform,Footnote 3 after which the transformed template is masked by a random filter. Image-based transformations are also applied towards protecting the finger vein template in [3], where block re-mapping and mesh warping are (separately) applied to the finger vein image to derive two versions of a cancellable finger vein template. Random projection is the template protection method of choice in [4], where the finger vein template consists of end points and intersections. Hybrid template protection strategies have been proposed for finger veins in [5, 6]. In [5], the finger vein image is first transformed into a template where the number of black (background) and white (vein) pixels is approximately equal, and then the Fuzzy Commitment scheme is applied to this template. In [6], the authors propose generating two BioHashes from the same finger vein template, then encrypting one BioHash using Fuzzy Commitment and the other using Fuzzy Vault, after which the two encrypted BioHashes are combined. Finally, [7,8,9] have focused on multi-biometric systems. More specifically, in [7], finger vein, fingerprint, finger knuckle print and finger shape features are fused, and then the resulting feature vector is secured via Fuzzy Commitment. A similar approach is presented in [8], except here the authors also consider score-level and decision-level fusion, whereby Fuzzy Commitment is used to secure each individual feature vector, then the scores or decisions, respectively, of the resulting biometric cryptosystems are fused. In [9], the finger vein feature vector is protected using the Bloom filter approach, and the authors also investigate a multi-biometric system whereby the Bloom filter-protected finger vein template is fused with a Bloom filter-protected face template.
This chapter contributes towards research on finger vein template protection by investigating whether the BioHashing template protection strategy [10] is suitable for protecting finger vein templates, in terms of its effect on the recognition performance of the underlying recognition system. BioHashing is one of the most widely studied biometric template protection schemes in the literature. It involves the projection of a biometric feature vector into a random subspace defined by a user-specific seed, followed by binarisation of the resulting projected vector to produce a so-called BioHash. Although BioHashing has been applied to a number of biometric characteristics (e.g. fingerprints [10], face [11], palm prints [12], and iris [13]), the only mention of BioHashing on finger vein templates that we have come across is the BioHashing/Fuzzy Vault and BioHashing/Fuzzy Commitment hybrid scheme in [6], mentioned earlier. To the best of our knowledge, there does not yet exist any published research on applying BioHashing on its own to finger vein templates. This is where our contribution lies. We also provide an open-source BioHash-protected finger vein verification system, which can be used by other researchers to verify and build upon our work.
We have chosen to focus on BioHashing for three main reasons. First, one of the biggest and most well-known advantages of BioHashing is that, theoretically, there is the possibility of achieving a 0% error rate. While low error rates may be characteristic of two-factor template protection schemes in general, BioHashing is currently the most popular in this category. Second, finger vein images tend to be fairly large, so we were interested in seeing whether BioHashing could be used to produce significantly smaller finger vein templates. Finally, since BioHashing is one of the most well-known template protection schemes in the literature, we wished to provide an open-source implementation of this method for comparison purposes against other template protection techniques developed for finger vein templates.
Note that the new standardFootnote 4 for the evaluation of biometric template protection schemes, ISO/IEC 30136:2018, specifies a number of requirements that should be considered when assessing the robustness of a biometric template protection scheme. These include the recognition performance of a biometric system employing template protection compared to that of the same system without template protection; the irreversibility of a template protection scheme, which refers to the difficulty of recovering information about the underlying biometric characteristic from its protected template; diversity, renewability (or cancellability), and unlinkability, all of which relate to the possibility of generating multiple protected templates from the same biometric characteristic, such that the protected templates are effectively seen as different identities and can thus be used to (i) replace a compromised protected template, and (ii) enroll into multiple applications using the same biometric characteristic without the risk of cross-matching the protected reference templates. The standard also specifies the need to evaluate the possibility of impersonating an enrolled individual using information about their underlying biometric characteristic leaked from one or more of their protected templates, which may largely be attributed to the template protection scheme’s compliance with the irreversibility and unlinkability properties. A thorough evaluation of a biometric template protection scheme must, therefore, take into account all of the aforementioned requirements. While the evaluation of recognition performance is relatively established, there are currently no solid, agreed-upon methods for assessing requirements such as irreversibility and diversity/cancellability/unlinkability (despite some guidelines provided by the new standard). Consequently, a thorough evaluation of a biometric template protection scheme necessitates a dedicated treatise of each requirement, which, in many cases, may involve the development and justification of new evaluation methodologies. In light of these reasons, this chapter focuses on evaluating only the recognition performance of BioHash-protected finger vein templates, and we reserve the analysis of the remaining requirements for future work.
The remainder of this chapter is structured as follows. Section 15.2 briefly describes the implementation of our BioHash-protected finger vein verification system. Section 15.3 presents experimental results on the recognition performance of this system and discusses memory constraints that should be considered when applying BioHashing to finger veins. Section 15.4 concludes the chapter and suggests areas for future work.
2 BioHash-Protected Finger Vein Verification System
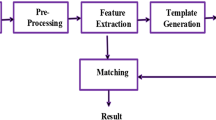
Our BioHash-protected finger vein verification systemFootnote 5 is an adaptation of the baseline finger vein verification system implemented in the PyPI package.Footnote 6 Our adapted system consists of four modules, as illustrated in Fig. 15.1.
The preprocessor locates, crops and horizontally aligns the finger in each finger vein image, as per [14, 15].
The extractor extracts the vein pattern from the cropped finger image. We used three well-known extractors: Wide Line Detector (WLD) [15], Repeated Line Tracking (RLT) [16] and Maximum Curvature (MC) [17]. The output of each extractor is a binary image, in which white pixels represent the finger vein pattern and black pixels represent the background. For each binary image, we then concatenate its rows to generate a finger vein feature vector.
The finger vein feature vector obtained from the feature extraction stage is next BioHashed. Our implementation is based on the original BioHash method proposed in [10]. The steps are summarised below:
-
1.
Generate a user-specificFootnote 7 random projection matrix of size \(n \times l\) for each unique fingerFootnote 8 in the database, where n represents the dimensionality of the finger vein feature vector and l denotes the desired BioHash length. To ensure that the same matrix can be generated for a specific finger during every verification attempt, the random matrix generation is seeded with a user-specific seed. (This seed should be stored on an external token, separately from the BioHash.)
-
2.
Orthonormalise the random matrix.
-
3.
Compute the dot product between the finger vein feature vector and each column of the orthonormalised random matrix. The result is an l-dimensional projected vector.
-
4.
Binarise the projected vector using the mean of the vector as the binarisation threshold, such that all values greater than the mean are set to 1 and all values less than or equal to the mean are set to 0. The result is an l-dimensional binary vector, referred to as the “BioHash”.
For the unprotected (without BioHashing) templates in our baseline finger vein verification system, comparison is performed on the extracted finger vein features separately for each of the three extractors (WLD, RLT and MC), using the comparison algorithm proposed in [16]. This method is based on a cross-correlation between the enrolled (reference) finger vein template and the probe template obtained during verification. For the protected (with BioHashing) templates in our BioHash-protected finger vein verification system, comparison is done by computing the Hamming distance between the reference and probe BioHashes.
3 Recognition Performance of BioHash-Protected Finger Vein Verification System
This section presents the results of the experiments we conducted to determine the recognition performance of our BioHash-protected finger vein verification system.
For the experiments reported in this paper, we employed the publicly available finger vein database UTFVP.Footnote 9 This database consists of four images for each of 60 subjects’ left and right index, ring and middle fingers, which makes up 1,440 images in total. Each image has a height of 380 pixels and a width of 672 pixels. Associated with the database are a number of different evaluation protocols. We used the “nom” protocol,Footnote 10 for which the database is split into three sets (“world”, “dev”, and “eval”). We employed the “eval” set, which consists of fingers 29–60. The comparison protocol involved using the first two finger vein images from each finger for enrolment and the last two as probes.
We chose this database for two reasons. First, it is publicly available, which means that our results can be easily verified by other researchers. Second, it has been shown [18] that an EER of as low as 0.4% is achievable on this database, so we wanted to investigate the effects of BioHashing on such remarkable recognition performance.
3.1 Baseline Recognition Performance
To determine how effective our BioHash-protected finger vein verification system is for finger verification purposes, it was necessary to first establish the recognition performance of our baseline verification system, i.e. using unprotected finger vein features. We had three baselines, one for each of the three extractors.
Figure 15.2 illustrates the outputs of each of the three feature extractors on a finger image from UTFVP, and Table 15.1 shows the dimensionalities of the finger vein feature vectors from each extractor. Although the images in Fig. 15.2 have all been scaled to the same size for easier visual comparison of the extracted patterns, the three extractors actually produce images of different sizes, as is evident from Table 15.1. The MC extractor is the only one that outputs a binary image of the same size as the original image from the database, plus a little extra background padding for comparison purposes. On the other hand, both the WLD and RLT extractors output binary images that are much smaller than the original image. Our adopted WLD extractor reduces the image to a quarter of its original size in each dimension prior to feature extraction to speed up the processing, and the RLT extractor reduces each dimension of the image to a third of its original size. These dimensionalities will be shown to play an important role in the practical feasibility of applying BioHashing to finger vein patterns, a point which will be discussed further in Sect. 15.3.3.
Figure 15.3 presents a visual comparison of the recognition performance of the three extractors in terms of Receiver Operating Characteristic (ROC) plots. We refer to this as the baseline recognition performance (i.e. the performance of the finger vein recognition systems prior to incorporating BioHashing).
Considering the recognition performance of the three extractor baselines in Fig. 15.3, it is evident that the MC extractor has the best performance. Looking at Fig. 15.2, this makes sense, because the MC extractor seems to produce the cleanest, thinnest finger vein patterns, which would be expected to contribute to more accurate recognition. The fact that the recognition performance of the WLD and RLT extractors is very similar may be attributed to the fact that the two extractors produce finger vein patterns of similar quality (thick, with a fairly noisy background), even though the RLT-extracted pattern in Fig. 15.2 appears cleaner than the WLD-extracted pattern.
3.2 BioHashing Recognition Performance
This section presents experimental results on the recognition performance of our BioHash-protected finger vein verification system. We consider two scenarios: the Normal scenario and the Stolen Token scenario. The Normal scenario refers to the scenario where each user of the verification system employs their own secret seed and associated random projection matrix in the generation of their BioHash. This is the expected scenario for most cases in practice. The Stolen Token scenario refers to the scenario where a genuine user’s secret seed is stolen and used with the impostor’s own finger vein template to generate the impostor’s BioHash. While it is hoped that such a scenario would not occur in practice, the fact that the user-specific seed is a valuable secret means that we must consider the scenario where that secret is leaked.
To determine the recognition performance of our BioHash-protected finger vein verification system in both the Normal and Stolen Token scenarios, we generated BioHashes of lengths \(l = \{100, 200, 300, 400, 500\}\) (number of bits) for finger vein feature vectors resulting from each of our three feature extractors (WLD, RLT and MC). For the Normal scenario, the unique ID of the finger image was used as the seed,Footnote 11 and for the Stolen Token scenario, the same seed (seed \(=\) 100) was used to generate the BioHashes for all fingers. Table 15.2 indicates the dimensionality reduction resulting from applying BioHashing to the finger vein feature vectors (refer to Table 15.1 for the original finger vein feature vector dimensionality). Figure 15.4 shows the recognition performance of the three finger vein extractors in both the Normal and Stolen Token scenarios, in terms of ROC plots.
From Table 15.2, it is evident that generating BioHashes of 100–500 bits from finger vein feature vectors results in a significant dimensionality reduction for all three feature extractors. The greatest dimensionality reduction is observed for the MC extractor, and the WLD extractor shows the smallest dimensionality reduction. This makes sense, since MC finger vein feature vectors have the largest dimensionality and WLD finger vein feature vectors the smallest (see Table 15.1). While “dimensionality” does not necessarily equal “information”, and thus “dimensionality reduction” does not necessarily imply “information loss”, the size of the dimensionality reductions noted in Table 15.2 makes it highly probable that mapping finger vein feature vectors to BioHashes does result in some information loss. In particular, from the results in Table 15.2, we would conclude that BioHashing on MC finger vein feature vectors would incur the largest information loss and WLD feature vectors the smallest. This should be evident when comparing the recognition performance of the BioHash-protected finger vein recognition system to the baseline system (i.e. the system without BioHashing). We refer to Fig. 15.4 for this purpose.
There a number of important observations from Fig. 15.4. First, in the Normal scenario, the BioHash-protected finger vein recognition performance for the WLD and RLT extractors is generally better than the baseline and has an error rate of approximately 0% at all FMR values, for \(l > 100\). This is interesting, since the BioHashes are significantly smaller than the original finger vein feature vectors, as noted in Table 15.2. However, the additional entropy introduced by the user-specific projection matrices makes the resulting BioHashes more discriminative than the original finger vein feature vectors, so the superior performance of BioHashes is not surprising. The fact that the BioHashed MC finger vein patterns struggle to reach the baseline recognition performance as quickly as WLD or RLT BioHashes is probably because BioHashing on MC finger vein feature vectors results in the largest dimensionality reduction (see Table 15.2). It is interesting to note, however, that although the dimensionality reduction for both RLT and MC is greater than 99% for all BioHash lengths tested (refer to Table 15.2), RLT BioHashes perform much better than MC BioHashes. So, perhaps such a large dimensionality reduction is too severe for MC finger vein patterns. Nevertheless, we can see that the recognition performance improves as the BioHash length increases, and for all three extractors, the Normal scenario recognition performance in the BioHashed domain equalises or surpasses the baseline recognition performance as the FMR approaches \(10^{-1}\).
As for the Stolen Token scenario, from Fig. 15.4 we can see that the recognition performance for all three extractors is significantly worse than the baseline. Such a trend has been shown for other biometric characteristics in the literature (e.g. [19]), and it makes sense because in the Stolen Token scenario we are essentially performing a huge dimensionality reduction using the same projection matrix for each finger.Footnote 12 So, here we see the “real” effect (i.e. without the additional entropy introduced by the user-specific projection matrix in the Normal scenario) of the significant dimensionality reduction reported in Table 15.2. Since we cannot, in general, expect better recognition performance than the baseline when the dimensionality of our feature vectors is reduced via random projection, the best we can hope for is that the performance of our BioHash-protected finger vein verification system in the Stolen Token scenario is as close as possible to our baseline. From Fig. 15.4, we can see that, as in the Normal scenario, the recognition performance in the Stolen Token scenario approaches that of the baseline as the BioHash length increases.
If we were to rank our three extractors in the Normal scenario based on Fig. 15.4, we would place WLD and RLT first equal, followed by MC. This is an interesting turn of events, since the baseline ranking in Fig. 15.3 is the opposite. Our suspicion is that this is due to the thinness of the finger veins extracted by MC, which means that the MC feature vector may need a much higher resolution than the WLD or RLT feature vectors. So, a BioHash in the range of 100–500 bits might just be too small to represent the MC features.
Ranking the three extractors in the Stolen Token scenario, once again MC takes last place, with WLD and RLT fighting for first. It seems as if WLD has slightly better recognition performance than RLT for all but a BioHash length of 500, where RLT marginally takes over. We would expect that the smallest feature vector, that produced by WLD, would incur the smallest information loss as a result of the smallest dimensionality reduction in the projection to a 100–500 bit BioHash, while the greatest information loss would be incurred by the largest feature vector, that produced by MC. So, we would predict that the WLD extractor recognition performance would be closest to its baseline and MC furthest from its baseline in the Stolen Token scenario. This is, more or less, what we observe in Fig. 15.4.
If we had to draw a conclusion about the suitability of applying BioHashing to a finger vein verification system based on the recognition performance observed in Fig. 15.4 alone, we would probably have to say that BioHashing is not a suitable template protection scheme in this case. While we would assume that the system would operate in the Normal scenario most of the time, in which case BioHashing would be great for achieving a 0% error rate with the WLD or RLT feature extractors (or even the MC extractor, depending on what FMR the system needs to operate at), unfortunately we cannot ignore the possibility of the Stolen Token scenario. Since the recognition performance of all three extractors in the Stolen Token scenario is significantly worse than the baseline for the BioHash lengths tested, it seems too risky to recommend incorporating BioHashing into a finger vein verification system.
However, we have observed that the recognition performance of the BioHash-protected finger vein verification system improves as the BioHash length increases. So, this brings to mind a possible solution: Why not just try larger lengths? We discuss this point in Sect. 15.3.3.
3.3 Memory Constraints
This section investigates the possibility of increasing the BioHash length to gain better recognition performance for our BioHash-protected finger vein verification system in the Stolen Token scenario. Since we know that, theoretically, we cannot achieve better recognition performance than the baseline in the Stolen Token scenario, our first approach might be to choose the MC extractor, since Fig. 15.3 shows that it has the best baseline out of the three extractors tested. Even though the recognition performance of the BioHashed MC finger vein features in Fig. 15.4 was shown to be worse than the performance of the WLD and RLT features, our hope might be that if we choose a large enough BioHash length then perhaps it would be possible to push the performance of our BioHashed MC features up to the MC baseline performance. The question is, how large would this BioHash need to be in order for us to achieve such an improvement in the recognition performance?
Figure 15.5 shows a plot of the amount of memory required, in bytes, to generate the projection matrix for a single feature vector for each of our three extractors, as the BioHash length increases from 100 to 2,000. Remember that the projection matrix consists of n rows by l columns, where n denotes the number of bits in the binary feature vector (see Table 15.1) and l represents the BioHash length.
From Fig. 15.5, we can see that the amount of memory required for a projection matrix corresponding to a WLD feature vector grows quite gradually as the BioHash length increases, that for an RLT feature vector grows faster, and that for an MC feature vector the fastest. For example, it seems that for a 1,000-bit BioHash we would require less than 0.1 GB for a WLD projection matrix, about 0.75 GB for RLT, and over 2 GB for MC! This immediately suggests that anything close to or larger than a 1,000-bit BioHash would probably be impractical for MC features, possibly doable for RLT features but not for a much larger l, and manageable for larger BioHashes on WLD features.
We attempted 1,000-bit BioHashes for our three extractors. As expected, the result was a memory error for our MC feature vectors (i.e. insufficient memory available). This confirms our suspicion that, although MC has the best baseline, it may be impractical for BioHashing. We might consider re-scaling the MC-extracted finger vein pattern image so that we have a smaller feature vector to work with, but this is currently not a characteristic of our adopted MC extractor implementation. As for the WLD and RLT extractors, Fig. 15.6 compares their recognition performance on 1,000-bit BioHashes in the Stolen Token scenario (note that both extractors had an error rate of 0% in the Normal scenario, so this is not shown).
As expected from the Stolen Token plots in Fig. 15.4, the recognition performance of the two extractors in Fig. 15.6 is fairly close, with RLT doing slightly better at the larger BioHash length. Overall, however, this recognition performance may still be impractically low, so we might need to consider an even larger BioHash length to try to improve the performance.
We attempted a BioHash length of 5,000 for our WLD and RLT features. As expected, the RLT-based BioHash generation resulted in a memory error. This means that, with our current implementation of the RLT extractor, we cannot expect to gain a significant improvement in the recognition performance of RLT-based BioHashes in the Stolen Token scenario. The WLD-based BioHashes, on the other hand, had no memory issues. Figure 15.7 compares the recognition performance of our BioHash-protected finger vein verification system for 1,000-bit and 5,000-bit BioHashes on the WLD finger vein features in the Stolen Token scenario to the WLD baseline (note that both BioHash lengths had an error rate of 0% in the Normal scenario, so this is not shown).
Figure 15.7 confirms our previously observed trend (in Fig. 15.4) that the recognition performance of our WLD-based BioHash-protected finger vein verification system approaches the performance of the corresponding baseline in the Stolen Token scenario as the BioHash length increases. The final length will depend on how much of a drop in recognition performance is acceptable in the Stolen Token scenario. Technically, we can expect the BioHash recognition performance to be approximately the same as the baseline performance when the BioHash length is the same as the length of the original feature vector. The issue here is that, in this case, the BioHash is more or less fully invertible, meaning that it would be possible to recover the original feature vector if the user’s secret seed and thus their projection matrix is leaked to an attacker. So, it is important to try to find a large enough BioHash length to ensure we have reasonable recognition performance in both the Normal and Stolen Token scenarios, while keeping the length small enough to ensure that the resulting BioHash is sufficiently privacy-preserving. The privacy-preserving properties of our BioHash-protected finger vein verification system must be investigated before we can fully justify any conclusions on whether or not BioHashing is a suitable template protection scheme for finger veins.
4 Conclusions and Future Work
This chapter presented the first investigation into the suitability of BioHashing as a finger vein template protection scheme for finger vein verification systems based on three feature extractors (WLD, RLT and MC), in terms of recognition performance only. Our experiments showed that, in the Normal scenario, it is possible to achieve a 0% error rate for BioHashes that are significantly smaller than the original finger vein feature vectors. BioHashes generated from WLD and RLT finger vein feature vectors were found to perform the best, while BioHashed MC features were shown to approach the baseline recognition performance as the FMR approached \(10^{-1}\). As expected, the recognition performance for all three extractors was worse than the baseline in the Stolen Token scenario due to the huge dimensionality reduction that is incurred in projecting a finger vein feature vector to a relatively small BioHash. While the recognition performance was shown to improve by increasing the length of the BioHash vectors, it was also demonstrated that the choice of length is constrained in practice by the amount of memory required for the projection matrix. Consequently, the WLD extractor was found to be the most promising for BioHashing purposes, since the relatively small size of WLD feature vectors allows for much larger BioHashes than would be possible for RLT or MC feature vectors. One issue with generating large BioHashes, however, is that, the larger the BioHash length, the easier it becomes to invert the BioHash to recover the original feature vector, thereby jeopardising the privacy of the verification system’s users. To determine an optimal BioHash length that would ensure a reasonable balance between recognition performance and privacy preservation, we would need to conduct a full security and privacy analysis for the BioHashed WLD finger vein patterns. This will form part of our future work. Another area for future work could be to investigate the effect on BioHashing recognition performance when the three extractors are modified to produce feature vectors of the same size.
Notes
- 1.
- 2.
- 3.
This is essentially the Fourier transform, constrained to a finite field.
- 4.
- 5.
Code available at the following link: https://gitlab.idiap.ch/bob/bob.chapter.fingerveins_biohashing.
- 6.
- 7.
Note that “user” refers to an individual using the finger vein verification system. While the standardised term would be “biometric data subject” or “individual”, we have chosen to retain the term “user” for consistency with [10].
- 8.
Each finger represents a different identity or “user”.
- 9.
- 10.
Defined by Idiap Research Institute. See https://www.beat-eu.org/platform/databases/utfvp/1/ for more details.
- 11.
In practice, the seed should be randomly generated. We only used the finger ID as the seed so that our results are more easily reproducible.
- 12.
Recall that each finger corresponds to a different identity.
References
Sandhya M, Prasad MVNK (2017) Biometric template protection: a systematic literature review of approaches and modalities. Springer International Publishing, Cham, pp 323–370
Hirata S, Takahashi K (2009) Cancelable biometrics with perfect secrecy for correlation-based matching. Springer, Berlin, Heidelberg, pp 868–878
Piciucco E, Maiorana E, Kauba C, Uhl A, Campisi P (2016) Cancelable biometrics for finger vein recognition. In: 2016 First international workshop on sensing, processing and learning for intelligent machines (SPLINE), July 2016, pp 1–5
Liu Y, Ling J, Liu Z, Shen J, Gao C (2017) Finger vein secure biometric template generation based on deep learning. Soft Comput 1–9
Favre M, Picard S, Bringer J, Chabanne H (2015) Balancing is the key: performing finger vein template protection using fuzzy commitment. In: 2015 International conference on information systems security and privacy (ICISSP), Feb 2015, pp 1–8
Yang W, Hu J, Wang S (2013) A finger-vein based cancellable bio-cryptosystem. Springer, Berlin, Heidelberg, pp 784–790
Lu L, Peng J (2014) Finger multi-biometric cryptosystem using feature-level fusion. Int J Signal Process Image Process Pattern Recognit 7(3):223–236
Peng J, Li Q, Abd El-Latif AA, Niu X (2014) Finger multibiometric cryptosystems: fusion strategy and template security. J Electron Imaging 23(2):023001
Gomez-Barrero M, Rathgeb C, Li G, Ramachandra R, Galbally J, Busch C (2018) Multi-biometric template protection based on bloom filters. Inf Fusion 42:37–50
Jin ATB, Ling DNC, Goh A (2004) Biohashing: two factor authentication featuring fingerprint data and tokenised random number. Pattern Recognit 37(11):2245–2255
Goh A, Ngo DCL (2003) Computation of cryptographic keys from face biometrics, pp 1–13. Springer, Berlin, Heidelberg
Connie T, Teoh A, Goh M, Ngo D (2005) Palmhashing: a novel approach for cancelable biometrics. Inf Process Lett 93(1):1–5
Chin CS, Jin ATB, Ling DNC (2006) High security iris verification system based on random secret integration. Comput Vis Image Underst 102(2):169–177
Lee EC, Lee HC, Park KR (2009) Finger vein recognition using minutia-based alignment and local binary pattern-based feature extraction. Int J Imaging Syst Technol 19(3):179–186
Huang B, Dai Y, Li R, Tang D, Li W (2010) Finger-vein authentication based on wide line detector and pattern normalization. In: 2010 20th International conference on pattern recognition, Aug 2010, pp 1269–1272
Miura N, Nagasaka A, Miyatake T (2004) Feature extraction of finger-vein patterns based on repeated line tracking and its application to personal identification. Mach Vis Appl 15(4):194–203
Miura N, Nagasaka A, Miyatake T (2007) Extraction of finger-vein patterns using maximum curvature points in image profiles. IEICE Trans Inf Syst 90(8):1185–1194
Ton BT, Veldhuis RNJ (2013) A high quality finger vascular pattern dataset collected using a custom designed capturing device. In: 2013 International conference on biometrics (ICB), June 2013, pp 1–5
Kong A, Cheung KH, Zhang D, Kamel M, You J (2006) An analysis of biohashing and its variants. Pattern Recognit 39(7):1359–1368
Acknowledgements
The authors would like to acknowledge the following sources of funding for supporting this work: the Secure Access Control over Wide Area Network (SWAN) project and the Swiss Center for Biometrics Research and Testing.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Krivokuća, V., Marcel, S. (2020). On the Recognition Performance of BioHash-Protected Finger Vein Templates. In: Uhl, A., Busch, C., Marcel, S., Veldhuis, R. (eds) Handbook of Vascular Biometrics. Advances in Computer Vision and Pattern Recognition. Springer, Cham. https://doi.org/10.1007/978-3-030-27731-4_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-27731-4_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27730-7
Online ISBN: 978-3-030-27731-4
eBook Packages: Computer ScienceComputer Science (R0)