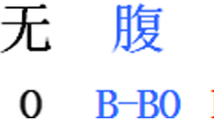Abstract
Clinical Named Entity Recognition for identifying sensitive information in clinical text, also known as Clinical De-identification, has long been critical task in medical intelligence. It aims at identifying various types of protected health information (PHI) from clinical text and then replace them with special tokens. Along with the development of deep learning technology, lots of neural-network-based methods have been proposed to deal with Named Entity Recognition. As one of the state-of-the-art methods to address this problem, Bi-LSTM-CRF has become the mainstream due to its simplicity and efficiency. In order to better represent the entity-related information expressed in the context of clinical text, we design a novel Capsule-LSTM network that is able to combine the great expressivity of capsule network with the sequential modeling capability of LSTM network. Experiments on 2014 i2b2 dataset show that the proposed method outperforms the baseline and thus reveal the effectiveness of the newly proposed Capsule-LSTM network.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
In clinical text, there are protected health information (PHI) such as name, phone numbers, occupation and location, etc. To protect these privacy information from disclosure, the Health Insurance Portability and Accountability Act (HIPAA)Footnote 1 promulgated in 1996 in the United States clearly stipulates that all medical text data in scientific research and business must be de-privacy processed first. To serve this purpose, the task of Clinical Named Entity Recognition (NER) is used to identify sensitive information, both the boundaries and semantic classes of target entities, and is known as Clinical De-identification.
In the early stage, NER systems for clinical purpose, such as MedLEE [1], SymText [2], MPlus [3], KnowledgeMap [4], HiTEX [5], cTAKES [6], and MetaMap [7], are rule-based. Later, machine learning based method become popular [8,9,10]. Among them Conditional Random Field (CRF) [11] finally takes the lead [12]. Up till now, CRF has been widely adopted as the final decoding layer for NER models, regardless of the underlying structure.
Frustratingly, Machine learning based method rely heavily on labour entensive feature engineering. However, along with the surge of deep learning technology, neural network approaches open a new way to the solution of NER and bring about lots of new state-of-the-arts [13,14,15,16].
Although great progress has been made in classical NER task, the application of NER system to the clinical problem have not been fully investigated, especially that of deep learning methods. As we go deeper into the problem, we find that many state-of-the-art methods appearing in traditional NER have not been fully investigated for clinical NER, especially Clinical De-identification. Actually, different from datasets in traditional NER task, clinical texts are highly formatted and entities appearing in different part of a clinical text can have different types even if they have the same surface form.
Finally, the main contribution of our study can be sumarized as follows:
-
Different from previous works that model texts in sentence-level, we move the first steps towards modeling texts in document-level in Clinical De-identification.
-
We designed a noval Capsule-LSTM network, which can combine the great expressivity of capsule network and the sequential modeling capability of LSTM network.
-
Experiments show that Capsule-LSTMs can outperform the original LSTMs in Clinical De-identification.
2 Related Work
2.1 Named Entity Recognition
Named Entity Recognition (NER) is an important task and has been extensively studied in the literature of Natural Language Processing, which aims at identifying named entities like person, location, organization, time, clinical procedure, biological protein, etc [17].
During the early stage, most of the approaches to NER have been characterized by the use of traditional statistical machine learning methods like Decision Tree [8], Maximum Entropy Model [18], Hidden Markov Model (HMM) [19], Conditional Random Field (CRF) [9], Supporting Vector Machine (SVM) [10] and Boosting Algorithm [20], etc. Approaches that fall into this category often require labour entensive feature engineering while also severely suffer from the data sparsity problem.
With the rapid development of deep learning technology, lots of neural-network-based methods have been proposed to address the task of Named Entity Recognition to reduce the feature engineering labour. Collobert et al. [21] proposed an effective neural language model for extracting text feature, which also tested on NER task by using a CNN-CRF architecture. Huang et al. [22] proposed a Bi-LSTM-CRF model that works well on NER task. Ma and Hovy et al. [23] and Santos et al. [24] successfully applied CNN over characters of words to incorporate character-level feature, whose outputs were then concatenated with the word-level embeddings. Chiu and Nichols et al. [13] presented a hybrid model of bi-directional LSTMs and CNNs that learns both character- and word-level features. Lample et al. [25] discarded word-level encoding and model sequence completely over character-level feature instead.
Later, Peters et al. [26], Rei et al. [27], Reimers and Gurevych et al. [28] and Yang et al. [29] either utilized external resources or applied multi-task learning paradigm. Yang et al. [14] systematically investigated the effect of combining discrete feature and continuous feature for a range of fundamental NLP tasks including NER. Cetoli et al. [30] incorporated prior knowledge of dependency relation between words and measure the impact of using dependency trees for entity classification. The benchmark of NER has been pushed to a new state-of-the-art.
More recently, Seyler et al. [31] performed a comprehensive study about the effect of the importance of external knowledge. Zhang et al. [15] introduced lattice LSTM to NER task and to alleviate the segmentation error in Chinese. Zukov Gregoric et al. [16] distributed the computation of a LSTM cell across multiple smaller LSTM cells to reduce the total number of parameters.
2.2 Clinical De-Identification for Privacy Protection
Clinical De-identification is very much like traditional NER and has been a hot topic in clinical natural language processing for a long time. The task of Clinical De-identification was first presented by Uzuner et al. [32], and require NER system to identify and anonymize protected health information that appears in clinical notes. The dataset they used was released as part of 2006 i2b2 event.
The history of Clinical De-identification is very similar with that of Name Entity Recognition, where there is also a shifting process from rule-based system to machine learning, and then to deep learning. In the earlier stage, almost all system for Clinical De-identification were based on machine learning [33]. Stubbs et al. [34] made a full reviews over automatic de-identification systems that appeared in 2014 i2b2 de-identification track, among which all systems are based on machine learning methods and many have used Conditional Random Field for inference.
Later, researchers are resorting to deep learning approaches such that large amount of human labour can be avoided. Wu et al. [35] developed a deep neural network to recognize clinical entities in Chinese clinical documents using the minimal feature engineering approach and outperform the previous state-of-the-art. Liu et al. [36] investigated the performance of Bi-LSTM-CRF with character-level encoding over clinical entity recognition and protected health information recognition.
However, different from traditional NER task, clinical texts are highly formatted and entities appearing in different part of a clinical text can have different types even if they have the same surface form. Up till now, the problem of Clinical De-identification is still far from being solved.
In our study, we try to tackle the problem in document-level, treating each document as an single instance. Then, we introduce a novel Capsule-LSTM network that combine both the expressivity of Capsule Network and the sequential modeling capability of LSTM network. And finally, to justify our methods, we have chosen the latest 2014 i2b2 dataset, which was distributed as part of the i2b2 2014 Cardiac Risk and Protected Health Information (PHI) tasks.
3 Proposed Approach
3.1 Overall Architecture
The basic model architecture follows the general structure of Bi-LSTM-CRF, which encodes sentences using conventional bi-directional long-short term memory (Bi-LSTM) network and model target label using conditional random field (CRF).
Practically, named entities are usually comprised of out-of-vocabulary words, which can greatly damage the performance of a NER system. Therefore, in addition to word embedding, we also incorporated a character-level Bi-LSTM for better representing out-of-vocabulary words, just as many previous works have done. (See Fig. 1)
The Architecture of our Bi-LSTM-CRF with character-level Bi-LSTM encoding. In the figure, we demonstrated how an input document \(\langle x_1,x_2,x_3,x_4,\dots ,x_T \rangle \) was encoded, and how named entity \(\langle x_3,x_4 \rangle \) of type TYPE1 can be identified via BIO tagging scheme, where \(x_3\) was labeled B-TYPE1 while \(x_4\) was labeled I-TYPE1.
Usually, there are two available tagging schemes, ‘BIOES’ or ‘BIO’, from which we prefer ‘BIO’ for its simplicity as it will incur less parameters to learn. Under the ‘BIO’ tagging scheme, an identified entity is defined as a sequence of words with the first word labeled with ‘B’ and any other trailing word labeled with ‘I’. As is shown in Fig. 1, The input document \(X=\langle x_1,x_2,x_3,x_4,\dots ,x_T \rangle \) with annotated entity \(\langle x_3,x_4 \rangle \) of type TYPE1 (TYPE1 is an entity type, such as NAME, PHONE, etc). Then the target label sequence is \(X=\langle y_1,y_2,y_3,y_4,\dots ,y_T \rangle \), where the target labels for the entity \(\langle x_3,x_4 \rangle \) is \(\langle y_3,y_4 \rangle \) with \(y_3=\text {B-TYPE1}\), which means \(x_3\) is the start of an entity of type TYPE1, and \(y_4=\text {I-TYPE1}\), which means \(x_4\) is an internal word of an entity of type TYPE1.
3.2 Long-short Term Memory
Long-short term memory (LSTM) was originally proposed by Hochreiter et al. [37] to deal with the gradient explosion and gradient vanishing problem of vanilla recurrent neural network, which consists of input gate \(i_t\), forget gate \(f_t\), output gate \(f_t\) and cell state \(c_t\). The computation of LSTM goes like Eq. 1.
Because of its powerful sequential modeling capability, LSTMs have been widely used for many natural language processing task including NER and achieved promising results.
3.3 Capsule Network
Initially proposed by Hinton et al. [38], capsule network divide vector representation into a number of capsules, or groups of neurons, and are able to better represent object in an image. It is assumed that each capsule may represent an entity that is present in the input, and neurons in the capsule may represent properties of this entity. Sabour et al. [39] apply capsule network to the task of MNIST digit classification and proposed the CapsNet that outperform previous state-of-the-art convolutional network by a large margin with the same number of parameters.
Typically, We use \(\varvec{u}_i\) to denote the i-th input capsule, \(\varvec{v}_j\) to denote the j output capsule, and \(W_{ij}\) as a bridging weight parameter between \(\varvec{u}_i\) and \(\varvec{v}_j\). The computation of CapsNet are mainly about routing, as is detailed in Algorithm 1, whose input \(\hat{\varvec{u}}_{j|i}\) can be obtained by \(\hat{\varvec{u}}_{j|i} = W_{ij}\varvec{u}_i\).

Following the intuition of CapsNet, we apply capsule network to NER, with the expectation that capsules inside are able to capture the information of named entities in clinical texts. More specifically, we use capsule network style computation inside LSTM, and propose a novel Capsule-LSTM.
3.4 Capsule-LSTM
The basic idea of Capsule-LSTM is to combine the great expressivity of capsule network and the sequential modeling capability of long-short term memory network.
To design such a structure, we begin by representing the cell state and the hidden state of LSTM as a groups of capsules. That is, \(h_t,c_t \in {\mathbb {R}}^{d_h}\) becomes \(H_t, C_t \in {\mathbb {R}}^{n_c \times d_c}\), where \(n_c\) is the number of capsules and \(d_c\) is the dimension of each capsule.
3.5 Training and Inference
To train our model, we follow Collobert et al. [21] to use sentence-level log-likelihood as objective function, shown in Eq. 3.
Under the convention of CRF, the label sequence probability can be rewritten as:
Here, \(D_{train}={(X_i, Y_i)}_{i=1}^{|D|}\) is our training set, \(\varTheta \) is our set of model parameters, \(\varPsi \) is the transition score between successive labels (documents are prepended with a \(\langle start \rangle \) label and appended with a \(\langle end \rangle \) label.), \(\varPhi \) is the emission score from word to label, and finally \(Z(X_i)\) is the normalization term associated with input \(X_i\). Just like the training of traditional CRF, we further add L1 and L2 regularization term to avoid overfitting. Therefore, the final loss function turns out to be:
where \(R(\varTheta )\) is the sum of L1 and L2 regularization term. During the training phase, we optimize our model against \(L(\varTheta )\) using Adam [40] algorithm with \(lr=0.005\), \(\beta _1=0.9\) and \(\beta _2=0.999\). And then in testing phase, we apply Viterbi algorithm to find out the label sequences with maximal probability for input documents.
4 Experimental Details
4.1 Dataset
Description. The dataset we used in our study is a corpus of longitudinal medical records, distributed as part of the i2b2 2014 Cardiac Risk and Protected Health Information (PHI) tasks, or 2014 i2b2 dataset for brevity. This dataset consists of 1304 medical records from 296 diabetic patients, and is officially splitted into training and testing set, where training set contains 790 documents while the testing set contains 514 documents.Footnote 2 Each document is a well-formatted medical record and named entities inside documents are annotated as text spans with corresponding entity types, where 22 entity types in total are concerned.
Data Preprocessing. To avoid the nuance of handling raw data, we resort to the publicly available i2b2toolsFootnote 3 that is developed based on the official evalution scripts of 2014 i2b2 challenge to load data. In this way, we convert raw data into conll format while keeping some formatting information such as end-of-line and indentation by introducing special tokens like \(\langle eol \rangle \) and \(\langle tab \rangle \). All number appearing in the data are replaced by the special token \(\langle num \rangle \). The Table 1 and Fig. 2 shows some basic statistics of this dataset after data preprocessing.
4.2 Model Comparison
Evaluation Metrics. The evaluation metrics used in our study is F1 score in SemEval’13 standard, which introduced four different ways (Strict/Exact/Partial/Type) to measure precision/recall/f1 results based on the metrics defined by MUC [41]. Following previous works, we evaluate models by measuring in Strict way, which counts entity matching on exact boundary match over the surface string, regardless of the type. In our experiments, we do not implement evaluating metrics by ourselves, but use the publicly available evaluation toolkit NER-EvaluationFootnote 4.
Model Settings. In our study, the following models were compared:
-
CRF. Traditional Conditional Random Field implemented by CRFsuite [12]. Feature template for this model is shown in Table 2.
-
Bi-LSTM-CRF. Use conventional Bi-LSTM network for both word- and character-level encoding, and CRF for target modeling.
-
Bi-Capsule-LSTM-CRF. Use Capsule-LSTM for word-level modeling, conventional Bi-LSTM for character-level modeling, and CRF for target modeling.
To make fair comparison, we use similar hyper-parameter settings across all of the above models, where character embedding dimension is 20, character-level LSTM size is 10, word embedding dimension is 50, word-level LSTM size is 100 and word context window size is 5. As for the newly proposed Capsule-LSTM, we set the number of capsules to be 25 and the dimension of each capsule to be 4. For all models, we pretrained word embeddings using Word2VecFootnote 5.
4.3 Results and Analysis
Overall Results. Table 3 shows the results on 2014 i2b2 dataset, whose F1 are reported (\(\pm 0.5\)) based on multiple runs. From this table, we can see that our newly proposed Bi-Capsule-LSTM-CRF outperform the Bi-LSTM-CRF baseline.
Document-level vs. Sentence-level. We compared the performance of all models in both document- and sentence-level. It is shown in Table 3 that models performs better under the document-level setting, when compared to that under the sentence-level setting, justifying our assumption that document-level context information makes a difference in recognizing entities in clinical text.
Ablation Study. For further insight into the effects of each module involved in Bi-Capsule-LSTM-CRF, we perform ablation analysis over our model under the document-level setting (Table 4).
5 Conclusion
In our study, we design a novel neural network structure called Capsule-LSTM, which combine the great expressivity of capsule network and the sequential modeling capability of long-short term memory network. Experiments over 2014 i2b2 dataset demonstrated the effectiveness of our model.
Notes
- 1.
- 2.
Here, we use the word document and the phrase medical records interleaved without distinction.
- 3.
- 4.
- 5.
References
Friedman, C., Alderson, P.O., Austin, J.H., Cimino, J.J., Johnson, S.B.: A general natural-language text processor for clinical radiology. J. Am. Med. Inform. Assoc. 1(2), 161–174 (1994)
Koehler, S.B.: Symtext: A Natural Language Understanding System for Encoding Free Text Medical Data. Ph.D. thesis (1998). AAI9829757
Christensen, L.M., Haug, P.J., Fiszman, M.: Mplus: a probabilistic medical language understanding system. In: Proceedings of the ACL-02 Workshop on Natural Language Processing in the Biomedical Domain. BioMed 2002, Stroudsburg, PA, USA, 2002, vol. 3, pp. 29–36. Association for Computational Linguistics (2002)
Denny, J.C., Irani, P.R., Wehbe, F.H., Smithers, J.D., Spickard, A. Rd.: The KnowledgeMap project: development of a concept-based medical school curriculum database. In: AMIA Annual Symposium Proceedings/AMIA Symposium. AMIA Symposium, vol. 2003, p. 195 (2003)
Zeng, Q.T., Goryachev, S., Weiss, S., Sordo, M., Murphy, S.N., Lazarus, R.: Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med. Inform. Decis. Mak. 6(1), 1–9 (2006)
Savova, G.K., et al.: Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. Jamia 17(5), 507 (2010)
Aronson, A.R., Lang, F.: An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. 17(3), 229–236 (2015)
Sekine, S., Grishman, R., Shinnou, H.: A decision tree method for finding and classifying names in Japanese texts. In: Proceeding Workshop on Very Large Corpra (1998)
Ratinov, L., Roth, D.: Design challenges and misconceptions in named entity recognition. In: CoNll 2009: Thirteenth Conference on Computational Natural Language Learning (2009)
Li, Y., Bontcheva, K., Cunningham, H.: SVM based learning system for information extraction. In: Winkler, J., Niranjan, M., Lawrence, N. (eds.) DSMML 2004. LNCS (LNAI), vol. 3635, pp. 319–339. Springer, Heidelberg (2005). https://doi.org/10.1007/11559887_19
Lafferty, J.D., McCallum, A., Pereira, F.C.N.: Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the Eighteenth International Conference on Machine Learning. ICML 2001, San Francisco, CA, USA, pp. 282–289. Morgan Kaufmann Publishers Inc. (2001)
Chiu, J.P.C., Nichols, E.: Named entity recognition with bidirectional LSTM-CNNs. Comput. Sci. 4, 357–370 (2016)
Yang, J., Teng, Z., Zhang, M., Zhang, Y.: Combining discrete and neural features for sequence labeling. In: Gelbukh, A. (ed.) CICLing 2016. LNCS, vol. 9623, pp. 140–154. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75477-2_9
Zhang Y., Yang, J.: Chinese NER using lattice LSTM. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia, July 2018, pp. 1554–1564. Association for Computational Linguistics (2018)
Gregoric, A.Z., Bachrach, Y., Coope S.: Named entity recognition with parallel recurrent neural networks. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Melbourne, Australia, pp. 69–74. Association for Computational Linguistics, July 2018
Yadav, V., Bethard, S.: A survey on recent advances in named entity recognition from deep learning models. In: Proceedings of the 27th International Conference on Computational Linguistics, Santa Fe, New Mexico, USA, pp. 2145–2158. Association for Computational Linguistics, August 2018
Chieu, H.L., Ng, H.T.: Named entity recognition: a maximum entropy approach using global information. In: COLING 2002: The 19th International Conference on Computational Linguistics (2002)
Zhou, G., Su, J.: Named entity recognition using an HMM-based chunk tagger. In: Proceedings of 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, Pennsylvania, USA, pp. 473–480. Association for Computational Linguistics, July 2002
Freund, Y., Schapire, R.E.: Experiments with a new boosting algorithm. In: International Conference on Machine Learning, pp. 148–156 (1996)
Collobert, R., Weston, J., Bottou, L., Karlen, M., Kavukcuoglu, K., Kuksa, P.P.: Natural language processing (almost) from scratch. CoRR, abs/1103.0398 (2011)
Huang, Z., Xu, W., Yu, K.: Bidirectional LSTM-CRF models for sequence tagging. CoRR, abs/1508.01991 (2015)
Ma, X., Hovy, E.: End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, pp. 1064–1074. Association for Computational Linguistics, August 2016
Santos, C.N., Guimarães, V.: Boosting named entity recognition with neural character embeddings. CoRR, abs/1505.05008 (2015)
Lample, G., Ballesteros, M., Subramanian, S., Kawakami, K., Dyer, C.: Neural architectures for named entity recognition. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego, California, pp. 260–270. Association for Computational Linguistics, June 2016
Peters, M., Ammar, W., Bhagavatula, C., Power, R.: Semi-supervised sequence tagging with bidirectional language models. In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, Canada, pp. 1756–1765. Association for Computational Linguistics, July 2017
Rei, M.: Semi-supervised multitask learning for sequence labeling. In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, Canada, pp. 2121–2130. Association for Computational Linguistics, July 2017
Reimers, N., Gurevych, I.: Reporting score distributions makes a difference: performance study of LSTM-networks for sequence tagging. In: Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, pp. 338–348. Association for Computational Linguistics, September 2017
Yang, J., Zhang, Y., Dong, F.: Neural word segmentation with rich pretraining. In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Vancouver, Canada, pp. 839–849. Association for Computational Linguistics, July 2017
Cetoli, A., Bragaglia, S., O’Harney, A., Sloan, M.: Graph convolutional networks for named entity recognition. In: Proceedings of the 16th International Workshop on Treebanks and Linguistic Theories, Prague, Czech Republic, pp. 37–45 (2017)
Seyler, D., Dembelova, T., Del Corro, L., Hoffart, J., Weikum, G.: A study of the importance of external knowledge in the named entity recognition task. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Melbourne, Australia, pp. 241–246. Association for Computational Linguistics, July 2018
Uzuner, Ö., Luo, Y., Szolovits, P.: Evaluating the state-of-the-art in automatic de-identification. J. Am. Med. Inform. Assoc. 14(5), 550–563 (2007)
Meystre, S.M., Friedlin, F.J., South, B.R., Shen, S., Samore, M.H.: Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med. Res. Methodol. 10(1), 70 (2010)
Stubbs, A., Kotfila, C., Uzuner, Ö.: Automated systems for the de-identification of longitudinal clinical narratives. J. Biomed. Inform. 58(S), S11–S19 (2015)
Wu, Y., Jiang, M., Lei, J., Xu, H.: Named entity recognition in Chinese clinical text using deep neural network. Stud. Health Technol. Inform. 216, 624–628 (2015)
Liu, Z., et al.: Entity recognition from clinical texts via recurrent neural network. BMC Med. Inform. Decis. Mak. 17(2), 67 (2017)
Hochreiter, S., Schmidhuber, J.: Long short-term memory. Neural Comput. 9(8), 1735–1780 (1997)
Hinton, G.E., Krizhevsky, A., Wang, S.D.: Transforming auto-encoders. In: Honkela, T., Duch, W., Girolami, M., Kaski, S. (eds.) ICANN 2011. LNCS, vol. 6791, pp. 44–51. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-21735-7_6
Sabour, S., Frosst, N., Hinton, G.E.: Dynamic routing between capsules. In: Guyon, I., et al. (eds.) Advances in Neural Information Processing Systems 30, pp. 3856–3866. Curran Associates Inc., New York (2017)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: 3rd International Conference on Learning Representations. ICLR 2015, San Diego, CA, USA, May 7–9, 2015, Conference Track Proceedings (2015)
Chinchor, N., Sundheim, B.: MUC-5 evaluation metrics. In: Fifth Message Understanding Conference (MUC-5): Proceedings of a Conference Held in Baltimore, Maryland, 25–27 August 1993
Acknowledgments
This work was partly supported by National Key Research and Development Program of China (2017YFB0802204), National Natural Science Foundation of China under U1636103, Grant 61632011, and 61876053, Key Technologies Research and Development Program of Shenzhen JSGG20170817140856618, Shenzhen Foundational Research Funding JCYJ20170307150024907.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, C., Li, J., Liu, Y., Du, J., Tang, B., Xu, R. (2019). Named Entity Recognition in Clinical Text Based on Capsule-LSTM for Privacy Protection. In: Wang, D., Zhang, LJ. (eds) Artificial Intelligence and Mobile Services – AIMS 2019. AIMS 2019. Lecture Notes in Computer Science(), vol 11516. Springer, Cham. https://doi.org/10.1007/978-3-030-23367-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-23367-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-23366-2
Online ISBN: 978-3-030-23367-9
eBook Packages: Computer ScienceComputer Science (R0)