Abstract
MicroRNAs (miRNAs) are small non-coding RNAs with a key role in the post-transcriptional gene expression regularization, thanks to their ability to link with the target mRNA through the complementary base pairing mechanism. Given their role, it is important to identify their targets and, to this purpose, different tools were proposed to solve this problem. However, their results can be very different, so the community is now moving toward the deployment of integration tools, which should be able to perform better than the single ones.
As Machine and Deep Learning algorithms are now in their popular years, we developed different classifiers from both areas to verify their ability to recognize possible miRNA-mRNA interactions and evaluated their performance, showing the potentialities and the limits that those algorithms have in this field.
Here, we apply two deep learning classifiers and three different machine learning models to two different miRNA-mRNA datasets, of predictions from 3 different tools: TargetScan, miRanda, and RNAhybrid. Although an experimental validation of the results is needed to better confirm the predictions, deep learning techniques achieved the best performance when the evaluation scores are taken into account.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
MicroRNAs are a particular family of RNAs characterized by a length of about 22 nucleotides originated from the non-coding RNAs [1]. Their ability to link with the target leads to mRNA degradation and the translation process’s block [2], preventing the production of proteins. These small molecules have an important role also in the control of many biological processes, of which homeostasis is one of the possible examples [3].
Since their first discovery in 1993 [4, 5], miRNAs are at the center of the scientific community’s interest. The computational analysis states that at last \(30\%\) of human genes are regulated by miRNA [6], and it has been shown that their dysfunction can lead to the development and progression of different kind of diseases, from cancer to cardiovascular dysfunction [7], and also neurological disorders such as Alzheimer’s [8]. Indeed, some of the works done so far describe the role of miR-21 in different kinds of diabetes mellitus [9] and refer to the miRNAs of the has-let-7 family as associated with metabolic disease [10]. Regarding cancer, miRNAs can act as tumor inducers or suppressors, therefore they are the core of the development of new drugs and therapeutic methods [11,12,13].
Given their importance, understanding the complex mechanisms behind the interactions between miRNAs and their targets is one of the challenges of these years. As a matter of fact, predicting which are the molecules that link together is of vital importance to produce a specific solution to their misbehaviour. That said, the use of biological approaches alone is not a sufficient strategy to resolve the problem because, as they only partially match, each miRNA has multiple mRNA targets and vice versa. Since there are more than 2000 miRNAs in the human genome [14], it is impossible to experimentally test the enormous number of possible combinations between miRNA and mRNA. It is time-consuming and costly at the same time. There is, therefore, a need to identify in advance miRNA-target interactions so to apply experimental approaches that can provide their functional characterization and, thus, the understand of their effects.
The bioinformatics community proposed many computational prediction tools which scope is to provide putative miRNA-target interactions to be evaluated in laboratory. The problem lies in the fact that those tool predictions are often inconsistent with each other. Indeed, the biological properties and parameters used in the algorithms are often different and not complete, so the scientist has difficulties in understanding which tool provides the best prediction and choosing the appropriate miRNA to validate. Up to now, only a limited number of targets have been experimentally validated as the tools suffer from a high level of false positives [15].
The tools available so far include different computational approaches mainly based on the modelling of physical interactions. However, new tools based on Machine Learning (ML) and Deep Learning (DL) are starting to emerge. These tools are able to automatically extract information from sequences: deepTarget [16] relies on autoencoders and Recurrent Neural Networks (RNN) to model RNA sequences and learn miRNA-mRNA interactions; miRAW [17] uses DL to analyse the mature miRNA transcript without making assumptions on which physical characteristics are the best suitable to impute targets. Other DL algorithms make use of selected features to predict the genes targeted by miRNAs; for example in [18] the authors used three conservative features, eight accessible features, and nine complementary ones to train a Convolutional Neural Network (CNN) classifier.
Many works have been done to compare the performance of the available tools, and for this we refer the reads to [15, 19,20,21,22,23,24,25]. Almost all of them compare the most famous tools: PITA, microRNA, miRSystem, miRmap, microT-CDS, CoMir, mirWalk, TargetScan, PicTar, miRU, RNAhybrid, miRanda etc. All these methods use specific features and parameters, but one of the main difference among them is that some generate original scores by which the interaction is evaluated (e.g. TargetScan), while others are based on the development of older tools, like microRNA, which is based on a development of the miRanda algorithm.
Despite their variety, only few tools are effectively used in standard procedures. One reason is the confidence of their results, and another one is the easiness that characterizes their use: the web-based service is the preferred platform, followed by the downloaded, and last are the packages. This and the fact that the targets identified by more than one tool are supposed to have a higher probability to be validated in the lab [22] made us choose to test integrated tools for the prediction of miRNA-mRNA interactions based only on selected features.
From the above-mentioned ones, we selected three tools: TargetScan, miRanda, and RNAhybrid. These are among the most used ones and generate scores which describe different interaction mechanisms that we believe are to be considered simultaneously. This selection gave us the possibility to use the knowledge available on the processes behind the miRNA-target interactions without redundancy. That is not true for those tools using multiple scores to describe the same feature or relying on a wide number of other tools since, if the scores are produced viewing the same characteristic, this could lead to a bias.
We tested miRNA-mRNA positive and negative interactions on the selected tools, their scores were collected, and a dataset was built. As reported in a previous study, the approach combining the results of different prediction tools achieves better performance than those obtained by the single ones [22, 35].
In the recent years, DL gained a huge success in many classification problems, outperforming ML models [26], so we decided to verify if a case study as the one we described could be resolved by DL architectures in a better way compared to ML methods.
In this work, we employ two datasets of predictions from 3 different miRNA-mRNA interaction tools, namely, TargetScan, miRanda, and RNAhybrid, to train two DL classifiers and three different ML models: support vector machine (SVM), logistic regression (LR), and random forest (RF). More precisely, we use a dataset of positive and negative miRNA-target interactions of small dimension (572 examples), and a larger one (13505 examples) obtained by generating new negative examples. As expected, from the results we observed that DL models need more time to train, but they achieve the best performance when the evaluation scores are taken into account. Given these results, we can confirm a limit of machine and deep learning: they both need a considerable amount of data to train on.
In Sect. 2 we briefly describe the main biological properties of the miRNA-mRNA interactions, the three selected tools used for the sequence-based prediction, the scores they produce, and we introduce the ML and DL algorithms we chose to test. In Sect. 3 we describe the data and the specifics of the DL and ML methods we trained. In Sect. 4 we compare their performance on the dataset, and in Sect. 5 we draw the conclusion of our study.
2 Background
2.1 miRNA-mRNA Interactions and Prediction Tools
As mentioned before, miRNAs have a key role in various biological processes, especially in gene expression regularization, by binding to mRNA molecules. The miRNA-mRNA interactions are predicted by computational tools that commonly evaluate four main features:
-
Seed region. The seed region of a miRNA is defined as the first 2 to 8 nucleotides starting from the \(5'\)-end to the \(3'\)-end which is the chemical orientation of a single strand of nucleic acid [27]. This is the small sequence where miRNAs link to their targets in a Watson-Crick (WC) match: adenosine (A) pairs with uracil (U), and guanine (G) pairs with cytosine (C). It is considered a perfect seed match if there are no gaps in the whole seed region (8 nucleotides) but other seed matches are also possible, like the 6mer where the WC pairing between the miRNA seed and mRNA is up to 6 nucleotides.
-
Site accessibility. The miRNA-mRNA interaction is possible only if the mRNA can be unfolded after its binding to the miRNA. The mRNA secondary structure can obstruct the hybridization, therefore the energy necessary to provide the target accessibility can be considered to evaluate the possibility that the mRNA is the real target of a miRNA.
-
Evolutionary conservation. A sequence conservation across species may provide evidence that a miRNA-target interaction is functional because it is being selected by positive natural selection. Conserved sequences are mainly the seed regions [28].
-
Free energy. Since the Gibbs free energy is a measure of the stability of a biological system, the complex miRNA-mRNA energy variation can be evaluated to predict which is the most likely interaction [29]: a low free energy corresponds to a high probability that the interaction will occur.
From the miRNA-target prediction tools that use the aforementioned features, three are particularly popular in the scientific community:
-
TargetScan. It was the first algorithm able to predict miRNA-target interactions in vertebrates. It has been upgraded several times in the years and now it estimates the cumulative weighted context score (CWCS) for each miRNA submitted. CWCS is the sum of the contribution of six features of site context to confirm the site efficacy [30], and it can vary from \(-1\) to 1. The lowest score is representative of a higher probability for the given mRNA to be targeted.
-
miRanda. It was also one of the earlier prediction tools. The inputs are sequences, and it searches for potential target regions in a sequence dataset. It outputs two different scores, one evaluating the alignment and the other the free energy: to describe a possible target, accordingly to the scores meaning, the former has to be positive and high, while the latter must be negative.
-
RNAhybrid. Given two sequences (miRNA and target), RNAhybrid determines the most energy favourable hybridization site. Thus, its main output is a score evaluating the free energy for a given seed region. The tool provides also a p-value score for the miRNA-mRNA interaction that is an abundance measure of the target site.
2.2 Machine Learning and Deep Learning
Given a miRNA and a mRNA we wanted to test if the combination of scores produced by different tools could give a more accurate indication of their likely to be linked (positive outcome) or not (negative outcome). This can be seen as a binary classification problem (true or false linkage) and, thus, ML and DL techniques are nowadays the most suitable choice to deal with this kind of question. They are automatic techniques for learning to make accurate predictions based on past observations [31]. The data used to learn the model represent the so-called training set, while the ones used to assess the generalization ability of the model is the test set. The learning performance is evaluated observing a chosen score, like the accuracy.
In the latest applications, DL approaches outperformed ML given their ability to learn complex features. This is the reason why we implemented DL architectures and compared their performance with suitable ML techniques like SVM, RF, and LR [33].
These methods have a very different characterization:
-
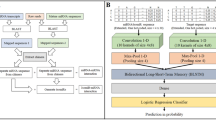
Deep Network. DL architectures are essentially neural networks with multiple layers which perform non-linear inputs elaboration [32]. A deep network is characterized by a large number of hidden layers, which relates to the depth. The number of layers is specific to the net because it indicates the complexity of the relationships it is able to learn. Another important parameter is the number of nodes in the layer. It is possible to choose between different kind of layers (e.g., dense, convolutional, probabilistic, or memory), each able to combine the input with a set of weights. A network with only dense layers is a standard DNN, a RNN is instead characterized by memory cells, like the LSTM. Non-linear functions like sigmoid and rectified linear unit are then used to compute the output. Deep networks are suitable to analyse high-dimensional data.
-
Support Vector Machines. SVMs are one of the most famous ML algorithms, capable of performing linear and nonlinear classification. They aim to select the coefficients for the optimal hyperplane able to separate the input variables into two classes (e.g., 0 and 1). SVMs perform better on complex but small or medium datasets.
-
Random Forest. A RF is an ensemble of decision trees. Decision trees are created to select suboptimal split points by introducing randomness. Each tree makes a prediction on the proposed data, and all the predictions are averaged to give a more accurate result. The ensemble has a similar bias but a lower variance than a single tree.
-
Logistic Regression. LR is the go-to method for binary classification in the ML area, commonly used to estimate the probability that an instance belongs to a particular class. The goal is to find the right coefficients that weight each input variable. The prediction for the output is transformed using a non-linear logistic function that transforms any value into the range 0 to 1. LR is commonly used for datasets that do not contain correlated attributes.
3 Methods
3.1 Data
The classifiers were trained with data from a reference dataset of 48121946 miRNA-target predictions. This dataset was obtained starting from the sequences of the miRNA families and of the untranslated regions (UTRs, the genomic loci targeted by miRNA) from 23-way alignment, filtering the information relative to the Homo sapiens species. More precisely, obtained a total of 30887 UTR (mRNA genes) and 1558 miRNA sequences, which were used as the starting point of our analysis. Then, we run the 3 tools (namely, miRanda, TargetScan, and RNAhybrid) and we combined their results in a matrix by looking at the positions on the UTR. Finally, to deal with the missing values of some tools, we assigned penalizing scores to them, which have been chosen after some experiments we conducted to assess how these penalizing scores influence the final classification results. As anticipated, the input matrix was composed of 48121946 rows (30887 UTRs \(\times \) 1558 miRNAs), in which the first five columns contain scores provided by TargetScan (Tscan-score), miRanda (miRanda-score and miRanda-energy) and RNAhybrid (RNAhybrid-mfe and RNAhybrid-pvalue). The last column contains the classification of the instances used to partition the dataset into five classes: negative examples (a), positive and experimentally validated examples (b), only experimentally validated examples (c), only positive examples (d) and unknown examples (e), as described in Table 1.
This latter classification is obtained by considering the predictions coming from [36] in which the authors generated two sets of “positive” and “negative” miRNA-target examples. The former set (positive examples) was obtained by biologically verified experiments, while the latter examples (negative) were identified from a pooled dataset of predicted miRNA-target pairs. Moreover, we downloaded the data from miRTarBase [37], a database of experimentally validated miRNA-target interactions, to additionally classify the positive examples into positive and experimentally validated examples, only experimentally validated examples, and only positive examples. More specifically, we crossed miRTarBase interactions with the positive examples from [36], in order to make the dataset more robust.
The unknown examples were not useful to train the classifiers, thus we put aside these examples, while we merged together b, c, and d data to obtain a unique positive class. The rearranged dataset was composed of 6841 positive and 286 negative interactions. A great class imbalance like the one present in this dataset is a huge problem for any ML or DL classification algorithm. In the training phase, the classifiers receive much more information of one class than of the other, and are not able to learn equally: the classifier tends to infer new data as part of the majority class. A possible way to deal with this problem is to make a balanced dataset by reducing the items of the majority class or by increasing the minority class examples. We tried both methods and compared the results obtained by the classifiers.
In the first case the dataset, that we called small dataset, was composed of the 286 available negative examples and 286 positive examples, sampled from the positive class: 179 from the original b classification, 54 from c, and 53 from d. In the second case, we constructed a dataset (called large dataset) of 13505 examples: 6664 positive and 6841 negative interactions. The negative examples were comprehensive of the 286 already available ones and 6555 new generated examples. The generation of the artificial negative examples was made through k-mer exchange between key and non-key regions of miRNA, as suggested in [34]. After their production, the miRNA were processed by HappyMirna [35], a tool for the integration of miRNA-target predictions and comparison. Thanks to HappyMirna we were able to obtain possible target for the new-generated miRNAs and the scores provided by TargetScan, miRanda, and RNAhybrid to construct a matrix equal to the previous one.
All datasets had missing values (NaN) whenever the prediction tools were not able to produce a result for the input. Since classifiers can not deal with NaN, we replaced all of them with penalizing scores chosen according to the range of the tool score, e.g. NaN for TargetScan were replaced with 1000. RNAhybrid was able to assign a score to 43744510 miRNA-mRNA interactions, while TargetScan only to 5341653 and miRanda to 4370618.
We observed that almost all the times a NaN was in the TargetScan record a corresponding NaN was also in miRanda. Instead, RNAhybrid does not fail to give a predictive value when the others meet a NaN.
3.2 Classifiers
We developed five classifiers (SVM, RF, LR, DNN, and RNN) using the python libraries Keras and Scikit-learn built on top of TensorFlow. All the classifiers were fine-tuned by selecting the optimal hyperparameters with GridSearchCV. When possible, we tried to maintain the same characteristics between models, as the number of folds for the cross-validation used during the training.
On the small dataset we trained the SVM, RF, LR, and DNN. The SVM best parameters were gamma = 1, C = 100, degree = 3, kernel = rbf, and random state = 100. The parameters tuned for the RF classifier were the number of estimators and the number of leaves for each estimator, obtaining 400 and 50, respectively. For the LR we investigated the solver, C, and the number of iterations, obtaining the best performance with the lbfgs solver, C = 3, and 50 iterations. For the DNN, we built a network with 5 fully connected layers, the first characterized by 80 nodes, from the second to the fourth with 40 nodes, and the last with 2 (necessary for a binary classification). The network was trained with 3000 epochs, a batch size of 100 and the ADAM optimizer. The score used to evaluate the performance was in all cases the accuracy. The training was with a 5-fold cross-validation.
On the large dataset, we used instead a 10-fold cross-validation. The best parameters for the SVM were gamma = 2, C = 100, degree = 3, kernel = rbf, and random state = 100. The RF had 100 estimators with a number of leaves for each estimator of 60. For the LR, C was set to 0.5, the iterations to 200, and the best solver was lbfgs. As for the deep architectures, we trained the same DNN used on the small dataset and a RNN characterized by three layers: the first and second were LSTM layers, with 150 and 100 units, respectively, the third was a dense layer like in the DNN. During the cross-validation, each fold was processed for 1000 epochs with a batch size of 1000.
4 Results
A way to compare the performance of different classifiers is to evaluate the accuracy, the ROC curve, and compute the area under the curve. Figure 1 shows the results obtained on the small dataset.
More precisely, the DNN and the RF had the best performance compared to the other classifiers, achieving an area under the curve of 0.66. Other measures were calculated and compared (see Table 2): the DNN had the best accuracy, precision, and f1 score over all the classifiers, while the recall was comparable. Based on this and the AUC we can state that the DNN was the model that best learned to recognize the miRNA-mRNA interactions.
All the positive data not used for the training were stored in separated datasets, called two and three dataset, so to remember the classification of origin. We used these datasets to check the percentage of the positive (and thus correct) predictions made by the trained classifiers, which was a way to further assess their performance. The same was done on the unknown data (the zero dataset) but in this case we could not say if the performance of the classifiers was good or not. In fact, since we did not have prior knowledge, we could only observe how different the predictions were. As we can see in Table 3, the SVM and DNN models were the ones able to recognize the higher number of interactions, especially on the three dataset. The RF model had the worst performance on all the datasets: we could not be sure of the results of this model as it gave only the \(50\%\) or less of correct evaluation on the unseen data that correspond to the two and three dataset. On the zero dataset the SVM and DNN were again the ones that proposed as possible the larger portion of interactions.
What we obtained training the models on the large dataset was instead very different. All the models were able to correctly classify over the \(95\%\) of the items, and none performed considerably better than the others (see Fig. 2 and Table 4). We believe that the scores improvement was thanks to the supplementary information the models were able to learn in the large dataset: miRNA-target interaction is a complex problem which needs a considerable amount of data to be addressed.
We trained the recurrent network to compare the results of the two deep architectures. Finding good parameters for the RNN was easier compared to the DNN, and also the number of necessary layers was smaller. Consequently, the training time was considerably reduced. As we used the three data and two data to build the large dataset, we could only observe how all the trained models behaved on the zero dataset (see Table 5): almost all the miRNA-target interactions were recognized as possibly true in all models.
5 Conclusions
miRNA-target interactions are predicted by a variety of tools that frequently give divergent results, thus are becoming diffuse solutions that integrate their outputs to give a unique and (possibly) reliable decision on the couples validity. Machine learning and deep learning methods are the ones preferably used to integrate different outcomes, with deep learning methods usually surpassing machine learning ones in terms of performance.
In our work, we trained five models from machine and deep learning area to test the possibility to identify a miRNA-mRNA interactions based on the scores provided by TargetScan, miRanda, and RNAhybrid. We used two different datasets: the first was a dataset of positive and negative miRNA-target interactions of small dimension (572 examples), the other was a larger dataset (13505 examples) obtained generating new negative examples. The performance of all the models was comparable in both cases: they performed poorly on the small dataset and very well on the large one.
Given these results, we can confirm a limit of machine and deep learning: they both need a considerable amount of data to train on. On the small dataset, comparing more scores, we said that the DNN performed fairly better than the other models, while on the large dataset we obtained from all the models very good and comparable results.
In the latter case we have to say that, as all the performance were very good (and as all the models gave the same results on the zero dataset), we do not suggest to use deep network solutions. In fact they have a difficult nature and they require a lot of time for the training and, moreover, on our problem the efforts did not give results upon the mean. It is much easier and faster to use standard machine learning implementations. On the small dataset the DNN results were better, but we can not recommend using this method over the others as the data were not enough to efficiently train the models.
In conclusion, we recognized the possibility to implement integrated tools based on machine and deep learning, the goodness of which can be finally evaluated only when the miRNA-target interactions they propose will be experimentally validated. The dimension of the dataset used to train is one of the main problems for a good integration. We showed that with a reduced dataset it’s hard to find a model which can easily recognized miRNA-target interaction (even if more DL architectures should be evaluated). If instead lots of examples are available, the choice of model type is irrelevant from the accuracy point of view.
References
Bartel, D.P.: MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297 (2004)
He, L., Hannon, G.J.: MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5(7), 522–531 (2004)
Liu, B., Li, J., Cairns, M.J.: Identifying miRNAs, targets and functions. Brief. Bioinform. 15(1), 1–19 (2012)
Lee, R.C., Feinbaum, R.L., Ambros, V.: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5), 843–854 (1993)
Wightman, B., Ha, I., Ruvkun, G.: Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75(5), 855–862 (1993)
Ross, J.S., Carlson, J.A., Brock, G.: miRNA: the new gene silencer. Am. J. Clin. Pathol. 128(5), 830–836 (2007)
Hackfort, B.T., Mishra, P.K.: Emerging role of hydrogen sulfide-microRNA crosstalk in cardiovascular diseases. Am. J. Physiol.-Heart Circ. Physiol. 310(7), H802–H812 (2016)
Hebert, S.S.: MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol. Dis. 33(3), 422–428 (2009)
Sekar, D., Venugopal, B., Sekar, P., Ramalingam, K.: Role of microRNA 21 in diabetes and associated/related diseases. Gene 582(1), 14–18 (2016)
Shi, C., et al.: Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 7(26), 40830–40845 (2016)
Ling, H., Fabbri, M., Calin, G.A.: MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 11, 847–865 (2013)
Samanta, S., et al.: MicroRNA: a new therapeutic strategy for cardiovascular diseases. Trends Cardiovasc. Med. 26(5), 407–419 (2016)
Riquelme, I., Letelier, P., Riffo-Campos, A.L., Brebi, P., Roa, J.: Emerging role of miRNAs in the drug resistance of gastric cancer. Int. J. Mol. Sci. 17(3), 424 (2016)
Hammond, S.M.: An overview of microRNAs. Adv. Drug Delivery Rev. 87, 3–14 (2015)
Akhtar, M.M., Micolucci, L., Islam, M.S., Olivieri, F., Procopio, A.D.: Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 44(1), 24–44 (2015)
Lee, B., Baek, J., Park, S., Yoon, S.: deepTarget: end-to-end learning framework for microRNA target prediction using deep recurrent neural networks. In: Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 434–442. ACM, October 2016
Planas, A.P., Zhong, X., Rayner, S.: miRAW: a deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PLoS Comput. Biol. 14(7), e1006185 (2017)
Cheng, S., Guo, M., Wang, C., Liu, X., Liu, Y., Wu, X.: MiRTDL: a deep learning approach for miRNA target prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 13(6), 1161–1169 (2016)
Bartel, D.P.: MicroRNAs: target recognition and regulatory functions. Cell 136(2), 215–233 (2009)
Mendes, N.D., Freitas, A.T., Sagot, M.F.: Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 37(8), 2419–2433 (2009)
Ruby, J.G., Stark, A., Johnston, W.K., Kellis, M., Bartel, D.P., Lai, E.C.: Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17(12), 1850–1864 (2007)
Alexiou, P., Maragkakis, M., Papadopoulos, G.L., Reczko, M., Hatzigeorgiou, A.G.: Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 25(23), 3049–3055 (2009)
Fan, X., Kurgan, L.: Comprehensive overview and assessment of computational prediction of microRNA targets in animals. Brief. Bioinform. 16(5), 780–794 (2014)
Srivastava, P.K., Moturu, T.R., Pandey, P., Baldwin, I.T., Pandey, S.P.: A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics 15(1), 348 (2014)
Faiza, M., Tanveer, K., Fatihi, S., Wang, Y., Raza, K.: Comprehensive overview and assessment of miRNA target prediction tools in human and drosophila melanogaster (2017). arXiv:1711.01632
Chen, H., Engkvist, O., Wang, Y., Olivecrona, M., Blaschke, T.: The rise of deep learning in drug discovery. Drug Discov. Today 23(6), 1241–1250 (2018)
Lewis, B.P., Burge, C.B., Bartel, D.P.: Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell 120(1), 15–20 (2005)
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B.: Prediction of mammalian MicroRNA targets. Cell 115(7), 787–798 (2003)
Yue, D., Liu, H., Huang, Y.: Survey of computational algorithms for MicroRNA target prediction. Curr. Genomics 10(7), 478–492 (2009)
Garcia, D.M., Baek, D., Shin, C., Bell, G.W., Grimson, A., Bartel, D.P.: Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18(10), 1139–1146 (2011)
Schapire, R.E.: The boosting approach to machine learning: an overview. In: Denison, D.D., Hansen, M.H., Holmes, C.C., Mallick, B., Yu.B. (eds.) Nonlinear Estimation and Classification. LNS, vol. 171, pp. 149–171. Springer, New York (2003). https://doi.org/10.1007/978-0-387-21579-2_9
Min, S., Lee, B., Yoon, S.: Deep learning in bioinformatics. Brief. Bioinform. 18(5), 851–869 (2017)
Goodfellow, I., Bengio, Y., Courville, A., Bengio, Y.: Deep Learning, vol. 1. MIT Press, Cambridge (2016)
Mitra, R., Bandyopadhyay, S.: Improvement of microRNA target prediction using an enhanced feature set: a machine learning approach. In: IEEE International Advance Computing Conference, pp. 428–433. IEEE, March 2009
Beretta, S., Giansanti, V., Maj, C., Castelli, M., Goncalves, I., Merelli, I.: HappyMirna: a library to integrate miRNA-target predictions using machine learning techniques. In: Proceedings of Intelligent Systems in Molecular Biology, July 2018
Bandyopadhyay, S., Mitra, R.: TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics 25(20), 2625–2631 (2009)
Hsu, S.D., et al.: miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 42, D78–D85 (2014)
Acknowledgments
This work was partially supported by national funds through FCT (Fundação para a Ciência e a Tecnologia) under project DSAIPA/DS/0022/2018 (GADgET).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Giansanti, V., Castelli, M., Beretta, S., Merelli, I. (2019). Comparing Deep and Machine Learning Approaches in Bioinformatics: A miRNA-Target Prediction Case Study. In: Rodrigues, J.M.F., et al. Computational Science – ICCS 2019. ICCS 2019. Lecture Notes in Computer Science(), vol 11538. Springer, Cham. https://doi.org/10.1007/978-3-030-22744-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-22744-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-22743-2
Online ISBN: 978-3-030-22744-9
eBook Packages: Computer ScienceComputer Science (R0)