Abstract
Here we describe preliminary results of a simulation-based study investigating the connection between tumor-treating fields (TTFields) distribution in the brain and glioblastoma patient outcomes. In order to perform this study, we developed a semiautomatic method for creating realistic head models from glioblastoma patient MRI using a deformable template and atlas-based registration. This method, which is described in detail in this chapter, is robust and fast, making it suitable for rapid creation of multiple realistic head models. Using this method, we created 119 head models of newly diagnosed glioblastoma patients that were treated with tumor-treating fields. Finite element simulations were used to simulate delivery of TTFields to these patients, and the connection between field intensity distribution at the tumor bed and patient outcome was analyzed. The result of this analysis support the hypothesis that increasing field intensity at the tumor bed improves patient outcome.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Tumor-treating fields (TTFields) are alternating electric fields in the intermediate frequency range (~100–500 kHz) known to exert an anti-mitotic effect on cancer cells [1,2,3]. The Optune™ device (Novocure, Ltd, Haifa, Israel) utilizes TTFields to treat glioblastoma multiforme (GBM). A pivotal clinical trial, EF-14, showed a significant benefit in overall survival in newly diagnosed GBM patients who received TTFields in addition to standard chemoradiation compared to patients who only received standard chemoradiation [4] (Fig. 7.1). The results of this trial led to the approval of the Optune™ device for the treatment of newly diagnosed GBM patients in multiple regions, including the USA, Canada, Europe, and Japan [5].
Components of the Optune™ device (bottom left) and model wearing the device (top left), as well as Kaplan-Meier curves showing overall survival for the treatment (blue line) and control (red line) groups of the EF-14 trial. The Optune™ device comprises a battery-operated field generator connected to four transducer arrays that are placed on the patient’s head. A backpack and carry-bag for the field generator as well as a battery charger and power supply are also shown in the photographs
The Optune™ device (see Fig. 7.1) is designed to deliver TTFields at a frequency of 200 kHz to the brain. 200 kHz coincides with the frequency at which the cytotoxic effect of TTFields on glioma cells is maximal [2]. Optune™ is a portable device comprising a battery-operated field generator, which is connected to transducer arrays through which TTFields are delivered (Fig. 7.1). Because TTFields affect cells dividing in parallel to the generated field more than other directions [1], Optune™ delivers TTFields in two orthogonal directions via two pairs of transducer arrays. The arrays are placed to deliver two electric fields in roughly orthogonal directions (see Fig. 7.1 (top left) and Fig. 7.4 (top row)). The device switches the field between the two sets of arrays once every second. The transducer arrays comprise nine circular disks each 1 cm in diameter. They are made from a ceramic with a relative dielectric constant >10,000. The disks make contact with the skin through a thin layer of conductive medical gel (~1 mm thickness). The disks are connected to one another using a flexible electric circuit (see bottom left in Fig. 7.1) and are geometrically arranged as shown in Fig. 7.4 (bottom row). The effect of TTFields is time dependent: the more time cells are exposed to the field, the stronger the effect. Therefore, to maximize the effect of treatment, patients are advised to maintain active therapy for at least 18 hr/day on average [6].
Preclinical results [1,2,3] have shown that the effect of TTFields is intensity dependent, and that the higher the intensity of the field, the stronger the cytotoxic effect of TTFields. The threshold intensity for observing the effect of TTFields is about 1 V/cm amplitude. Several simulation-based studies have shown that it is possible to maximize field intensity in the tumor by carefully selecting the position of the arrays on the scalp [7, 8]. Indeed, the NovoTAL™ system is a software-based system that utilizes morphometric measurements of head size, tumor size, and position (which are determined from a patient MRI) in order to optimize the position of the arrays on the head [9].
The dose-dependent nature of TTFields has been established in preclinical studies [1,2,3]. However, in order to fully understand the effect of dose and develop effective treatment planning strategies, it is important to establish the connection between TTFields distribution in the brain and patient outcome in a rigorous manner. Studies addressing this connection require estimating TTFields distribution within a large cohort of patients treated with TTFields over the course of their disease. Since physical measurement of the field distribution is highly invasive, and therefore challenging, numerical simulations utilizing realistic computational models of actual patients are the only practical means for performing such studies.
In order to perform such a study, two challenges need to be addressed:
-
1.
The availability of a dataset encompassing a large number of patients treated with TTFields. The dataset needs to include imaging data from which realistic head models of patients, including the tumor(s), can be obtained, and records of patient outcome including progression and survival.
-
2.
Development of a method for creating realistic computational patients in a robust and rapid manner.
The EF-14 clinical trial dataset includes detailed clinical data on over 400 patients treated with TTFields. Therefore, this dataset is well suited for this study. However, estimating TTFields distribution within these patients requires algorithms that enable construction of realistic head models of patients from MRI scans in a rapid and robust manner. Various pipelines have been adapted for the purpose of simulating the delivery of TTFields to realistic head models. Wenger et al. [8] utilized a pipeline that relied on FSL FLIRT [9], SimNibs [10], and Brainsuite [11] in order to create a realistic head model of a healthy individual into which artificial tumors of various shapes were inserted. Korsheoj et al. [12, 13] presented a different pipeline, utilizing SimNibs to create realistic head models of cancer patients. A different approach was presented by Timmons et al. [14], who utilized SPM8 [15] and ScanIP [16] to create realistic patient models. All of these approaches require a significant amount of human intervention and are therefore time-consuming and not suitable for a study requiring the creation of a large number of head models. An alternative approach could be to estimate the tissue conductivity from imaging data. Indeed, diffusion tensor imaging (DTI) [17] or alternatively water content-based electric property tomography (wEPT) [see Chap. 20 on wEPT in this book] could potentially be used to create realistic head models. However, both these sequences require specifically-adapted image series not available for all patients in the EF-14 study.
To overcome these challenges, we developed a novel method for creating realistic head models of patients utilizing a model of a healthy individual which serves as a deformable template. In this chapter, we present a detailed description of our method and demonstrate its utility by investigating the connection between TTFields field distribution and patient outcome in 119 patients treated with TTFields as part of the EF-14 trial.
2 Methods
2.1 MRI Data Used for the Study
MRI datasets analyzed in this study were obtained from the patient records of the EF-14 trial participants. This trial was a multi-center, open-label, randomized clinical phase 3 trial, which recruited 695 patients at 83 sites. Patients were randomized at the end of radiotherapy at a ratio of 2:1 to receive standard maintenance temozolomide chemotherapy with or without the addition of TTFields. To create patient models, T1-postcontrast MRIs at baseline (postsurgery and postradiation therapy) of 119 patients from the treatment arm of the trial were selected. Only patients who received TTFields therapy for over 2 months were selected. In general, for all patients, the baseline data contained T1-postcontrast data acquired from at least two of the possible three orientations (axial, sagittal, and coronal).
2.2 Image Preprocessing
Patient data was retrieved from the trial records in DICOM format. The DICOM data was imported and converted to NIfTI file format. The header of the NIfTI files was manipulated so that the origin of the file matched the origin of the template tissue probability maps (which are described in Sect. 7.2.6). This step ensures that the MRI images can be registered into the deformable template space. The NIfTI data was padded to add margins to the 3D image and resliced to a uniform grid of 1 × 1 × 1 mm.
2.3 MRI Full Head Completion
In order to create a head model from MRI data, it is important that the field of view of the MRI image show the entire head. However, in the clinic, where the focus is to image the tumor, the field of view of the full image set does not always show the entire head. Therefore, in cases where a single T1-contrast image showing the entire head was not available, we combined T1-contrast images acquired at different orientations in order to complete the field of view. In many cases, the image set acquired at one orientation had higher quality than the images acquired at other orientations. Therefore, the image with the highest quality was used as an anchor, and the other images were rigidly registered to it, followed by a histogram matching of the additional images to the anchor image. In order to create a new image, all voxels in which the original image contained MRI data were assigned the same value as the original image in the corresponding voxel. In the area of missing data of the anchor image (outside the borders of the original MRI series), the value of the voxels were set as the average of all nonzero values of the additional images. Figure 7.2 shows an example of an image set to which this algorithm was applied along with an output image in which the whole head is visible.
Example of an image set for which the MRI head completion algorithm was applied. For this patient, T1-postcontrast image datasets were captured at axial, coronal, and sagittal orientations existed (first three columns). However, the fields of view in all three image sets did not cover the entire head. An image depicting the full head was created by combining these image datasets using the field of view completion algorithm (column 4). Column 5 shows an overlay of the final patient model on the image of the complete head
2.4 High-Resolution Reconstruction
The original T1-contrast MRI data is often of lower resolution, which affects the tumor segmentation quality and the accuracy of the head model. A super-resolution algorithm that combines several T1-contrast images of the patient acquired at different orientations into a single high-resolution image was implemented. Based on [18], the best-quality image was set as the anchor image, followed by affine registration of the additional images to it. All images were resliced to a uniform grid using trilinear interpolation and a gray-scale intensity normalization was performed. The value of the voxel in the reconstructed image was a weighted average of the values of the corresponding voxels in the images used for reconstruction. When averaging, a higher weight was given to voxels from slices that were present in an original image versus voxels originating from slices obtained through interpolation.
2.5 Background Noise Reduction
A thresholding method was used to remove background noise and aliasing, both of which were found to deteriorate the quality of the head model created using deformable templates. In particular, when background noise was present, the contour of the skull obtained during model creation was often inaccurate and included part of the background. The background noise reduction was performed using a semi-automatic method in which the user selected a single value representing the background noise and the software applied this value as a threshold to automatically detect the contour of the scalp in the MRI image and set the intensity of the background to zero.
2.6 Patient Model Creation
Figure 7.3 is a schematic describing the pipeline used to create head models of glioblastoma patients using a deformable template. A prerequisite for this technique is to create a realistic head model of a healthy individual. In order to create a patient model from MRI images, the user first segments the tumor using manual or semiautomatic procedures. The region of the tumor is masked, and a nonrigid registration algorithm is used to register the MRIs to the template space, yielding a transformation from the patient space to the template space. The inverse transformation is then calculated and applied to the deformable template to yield an approximation of the patient head in absence of the tumor. Finally, the tumor is transplanted into the deformed template model to yield the final patient model.
Schematic showing the process used to create patient-specific models from a deformable template. A prerequisite is the creation of a realistic head model of a healthy individual, which serves as the template. To create a patient model, first, the tumor is manually segmented using ITK-SNAP. The segmented region is masked, and the masked MRI registered into the template space using a nonrigid registration algorithm. The registration yields a transformation from patient space to template space as well as an inverse transformation, which is applied to deform the template into the patient space. This yields a model which resembles the patient brain in the absence of a tumor. Finally, the tumor is implanted into the deformed head model to yield the final patient model
To reduce the schematic to practice, first a deformable template needs to be created:
The deformable template is represented using tissue probability maps (TPMs), which are a set of six 3D matrices that assign to each voxel a probability of belonging to a predefined tissue (white matter, gray matter, CSF, skull, scalp, and air). For the creation of the deformable template, the MRI of a healthy male based in MNI space [19] was segmented as TPMs. The procedure was performed using an algorithm that simultaneously registers and segments the base MRI using an existing set of TPMs (built in a standard space) and applied using the MATLAB toolbox SPM8 [15] and its extension MARS [20]. Manual corrections were made to the deformable template TPMs by manipulating the combination of probabilities in a specific voxel. Manual corrections included mainly adjustments to the regions of skull and scalp, so that a better match between the deformable template and patient MRI data was obtained in these the regions. A final step in creating deformable template TPMs from these probability maps is to apply a smoothing filter to the individual maps. Smoothing is important to allow adjustments to an MRI of any individual. The smoothing was performed using a Gaussian filter with a smoothing kernel of 4 × 4 × 4 mm FWHM (full width half maximum). It is noteworthy that the TPMs generated using this procedure is sharper than the original atlas TPMs of SPM/MARS. This method ensures that the segmented model has very high resemblance to the patient’s MRI data. Deformable template TPMs creation is performed once for the entire database.
T1-postcontrast MRIs were used to create the patient models. Pre-processing operations were performed when image quality or image resolution were insufficient, as described in Sects. 7.2.2, 7.2.3, 7.2.4, and 7.2.5. Following preprocessing, tumors and abnormal tissue were segmented in a semiautomatic manner using ITK-SNAP [21]. The following tumor and abnormal tissue types were segmented: enhancing tumor, necrotic core, enhancing nontumor (coincides with scarring), resection cavity, and skull defects. Furthermore, the following abnormal tissue regions were contoured/segmented: hematoma, ischemia, atrophy, and non-GBM tumor. Abnormal tissue regions were masked in the patient MRI, and the masked images were registered into MNI space using MARS/SPM and the TPMs created from the deformable template. This registration process yielded a nonrigid transformation from the patient space into the template space as well as the inverse transformation from template space to the patient space. The inverse transformation was applied to the probability map for each tissue type independently. A model approximating the patient in the absence of a tumor was then created by assigning to each voxel the tissue type for which the probability value within the voxel was highest. Finally, the manually segmented abnormal tissues were inserted to yield the final patient model.
2.7 Placement of Transducer Arrays on the Model
Optune™ transducer arrays comprise a set of nine ceramic disks, which make contact with the skin through a thin layer of medical gel. The disks are arranged in a well-defined geometry as shown in Fig. 7.4. Treatment planning to determine the optimal layout of the transducer arrays on the patient’s scalp is performed prior to beginning TTFields therapy. Treatment planning for patients that received TTFields as part of the EF-14 trial was performed. The treatment plan (optimal layout) for each patient was recorded in the patient’s clinical record. When simulating delivery of TTFields to a specific patient, arrays were placed on the model in a manner that matched the treatment plan saved in the patient record.
Example showing an array layout as stored in the patient record (top row), along with a patient model onto which arrays matching this layout have been placed (bottom row). Each array layout comprises four transducer arrays, shown as patches of blue, red, yellow, and white in the top row. The arrays are arranged into pairs and electric fields generated between each pair. One pair of arrays delivers an electric field with an anterior-posterior orientation (red and blue arrays in top row), and one pair delivers a field oriented left-right on the patient (white and yellow arrays in top row) Each transducer array comprises nine ceramic disks arranged in a rectangular orientation Each red disk on the model in the bottom row represents a ceramic disk. The bottom row shows a model onto which a pair of transducer arrays has been placed. One array was placed on the left aspect of the model, and the other on the right aspect
It is important to note that the optimal array layout for a patient was selected from a library of predefined layouts, an example of which can be seen in Fig. 7.4. The position of each array in each layout can be demarcated relative to well-defined anatomical landmarks. In addition, the arrays are constructed from nine discrete disks, set out in a well-defined and rigid geometry. These two observations led to the following procedure for placing the virtual arrays on the patient models.
2.7.1 Automatic Identification of Landmarks and Determination of the Array Positions
In order to determine the location and orientation of the disk array on the head, it is important to identify landmarks, as well as rotate the head model to a well-defined orientation relative to which the orientation of the array is known. An iterative algorithm detecting the orientation of the head and the landmarks was therefore used. The anatomical landmarks that were automatically identified were the centers of the eyes, brainstem, frontal sinuses, and the box bounding the head. Detection of landmarks was performed on the segmented model. The first step was to find the two CSF circles of the eyes in 2D axial slices. For each eye, the slice in which the area of the circle was maximal was found and its center identified. These two points correspond to the centers of the eyes. Using the center of the eyes, an initial correction of the head orientation was performed. The next step was to detect the brainstem (segmented as a round region of white matter in the base of the brain), and to perform full rotation of the model to a predetermined orientation. Once the orientation was completed, the frontal sinuses (segmented as air), and bounding box were found. These landmarks were then used to define the positions of the central disks of each array on the scalp as planned for each patient by the NovoTAL™ system.
2.7.2 Positioning of Anchor Points to Assist with Array Placement
After finding the position of each central disk of an array on the scalp, four anchor points at predetermined distances from the central disk were located on the scalp to assist with placement of the remaining disks. The anchor points were positioned to form a cross-like pattern with the central disk of the array at the center. One axis of the cross was vertically oriented, and the second axis of the cross was horizontally oriented. Each anchor point was found using the following iterative algorithm:
-
(a)
Calculate the tangent surface to the scalp at the current point using singular value decomposition (SVD).
-
(b)
Project the vector pointing in the direction of the anchor point onto the tangent surface.
-
(c)
Calculate a point at a predefined (small) distance from the current point along the direction defined by the vector calculated in step (b).
-
(d)
Define a new point on the scalp, as the point on the scalp closest to point calculated in step (c).
-
(e)
If the geodesic distance between the new point and the center point is close enough to the desired distance between center of the central disk and the anchor point, then set the current point as the anchor point, else perform another step towards the anchor point by repeating steps a–e.
2.7.3 Finding the Center of All Disks in an Array
The geometry of the transducer array suggests that the geodesic distances between the centers of the disks are constrained and remain constant when the array is placed on the scalp. Consequently, in order to find the center of a specific disk in the array, geodesic circles with appropriate radii were drawn around the central disk and the anchor points. The approximate intersection of these circles (defined as the point at which the sum of distances from the circles is minimum) corresponds to the center of that specific disk.
2.7.4 Creating Cylinders Representing the Ceramic Disks and the Medical Gel
When placing the arrays on the head, the ceramic disks are tangent to the scalp. The following process was used to ensure that the virtual disks are tangent to the scalp in the patient model. First, the normal direction to the body closest to the disk was calculated. The calculation was performed by finding all points on the phantom skin that were within a distance of one disk radius from the designated point. The coordinates of these points were arranged into the columns of a matrix and SVD performed on the matrix. The normal to the surface is the eigenvector that corresponds to the smallest eigenvalue. To ensure good contact between the disk and the body, the thickness of the gel needs to be in contact with all points under the disk. This was determined by fitting a cylinder to all points on the skin under the disk. Calculation of the positions, orientations, and gel thicknesses associated with the disks was performed in MATLAB 2013b (Mathworks, USA) and the Sim4Life (ZMT-Zurich, Switzerland) Python API was used to generate the transducer arrays in the model.
2.8 Simulations
Following creation of the patient models, transducer arrays were placed on the models to match the transducer array layouts assigned to the patients as recorded in their medical records. In addition, patient’s average compliance (defined as the fraction of time a patient was on active treatment) and the average electrical current delivered to each patient were calculated from log files of the TTFields generators stored in the patient records. The Sim4Life (ZMT Zurich, Switzerland) quasi-electrostatic solver was used to simulate the delivery of TTFields at 200 kHz. Electrical properties were assigned to the various tissue types and materials in the model according to average values reported in the literature [7, 17]. Boundary conditions were set so that the total current delivered to the patient was equal to the average current delivered to the patient during the first 6 months of treatment. It is important to note that a separate simulation was performed for each of the two pairs of arrays placed on the patient.
2.9 Analysis
For this study, a total of n = 119 cases (of 466 patients in the trial treatment arm) were simulated. All patients analyzed in this study were on active treatment for more than 2 months. For each patient, field intensity distributions in a tumor bed comprising the gross tumor volume (GTV) and a proximal boundary zone (PBZ) extending 1 cm from the GTV were derived. To account for compliance, field values for each patient were multiplied by their average compliance over the first 6 months of treatment.
To test the hypothesis that patient outcome correlates with field intensities, two quantities were derived:
-
1.
E95, which is the value such that 95% of the combined volume of the GTV and PBZ, received field intensities (multiplied by compliance) above a specified value.
-
2.
Eaverage, which is the average intensity (multiplied by compliance and average current) in the combined volume of the GTV and PBZ.
Patients were divided into two groups based on threshold values of both E95 and Eaverage, and the overall survival (OS) and progression-free survival (PFS) of the groups were compared. The threshold values of these parameters were chosen to yield the most statistically significant difference in OS between the groups.
3 Results
Demographics of all groups were similar to the demographics of the entire EF-14 trial population and similar to each other (Table 7.1). When dividing the 119 patients into two groups based on E95, the median OS and PFS were superior when E95 > 1.3 V/cm: OS (E95 > 1.3 V/cm: 33.0 months vs. E95 < 1.3 V/cm:21.9 months, p = 0.009, HR = 0.46) and PFS (E9 > 1.3 V/cm 11.9 months vs E95 < 1.3 V/cm 7.5 months, p = 0.06, HR = 0.49). Similar results were seen when dividing the patients into two groups based on a threshold of Eaverage > 1.0 V/cm: OS (Eaverage > 1.0 V/cm: 26.1 months vs. Eaverage < 1.0 V/cm: 21.6 months, p = 0.025, HR = 0.50) and PFS (Eaverage > 1.0 V/cm: 9.9 months vs. Eaverage < 1.0 V/cm: 6.1 months, p = 0.026, HR = 0.54). Progression-free at 6 months and 24-months survival rate were superior when E95 > 1.3 V/cm (E95 > 1.3 V/cm: 76% vs. E95 < 1.3 V/cm: 56%, p = 0.031), and (E95 > 1.3 V/cm: 66% vs. E95 < 1.3 V/cm: 42%, p = 0.0156), respectively. Within the E95 > 1.3 V/cm group, a higher percentage of patients showed clinical benefit (stable disease/partial response, 97% vs. 83% p = 0.039). OS in the E95 < 1.3 V/cm group was superior to OS in the EF-14-control-arm (16.0 months, p = 0.009), indicating that patients benefited from treatment even when field intensities were lower. The Kaplan-Meier curves for OS are shown in Fig. 7.5.
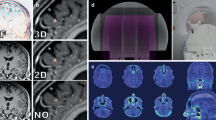
(i) Examples of patient models and calculated field distributions within each patient head. Each row shows a different patient. The figure shows axial slices from patient MRI (1st column), an overlay of the patient model on the slice (2nd column) and the final patient model in the same slice (3rd column), 3D representation of the patient model with virtual arrays (4th column) and field intensity distributions generated by the pair of arrays on the left and right aspects of the head (5th column) and on the anterior and posterior of the head (6th column). (ii) Kaplan-Meier curves for overall survival when splitting patients according to threshold values of Eaverage (left) and E95 (right)
4 Discussion and Conclusion
In this work, we summarize a simulation-based study suggesting a strong connection between TTFields dose at the tumor bed and patient outcome. A robust and rapid semiautomatic procedure for creating realistic patient models was developed in order to perform this study. This method enabled the efficient creation of patient-specific models and simulation of TTFields delivery to over 100 patients. Simulating delivery of TTFields to such a large cohort is necessary in order to gain a sample size large enough to establish statistically significant connections between dose/field intensity and patient outcome. To our knowledge, this is the first study in which delivery of low-frequency electromagnetic energy to patients has been performed on such a large cohort. In the future, our modeling process could be adapted to investigate potential connections between electromagnetic field distributions and patient outcomes for other electrotherapeutics. A study utilizing this method to analyze the connection between TTFields dose distribution and patient outcome in 340 patients that participated in the e EF-14 trial was recently published [22].
The head model creation procedure used in this study utilizes a healthy head model, which serves as a deformable template from which the patient models are created. Consequently, the anatomy of the resulting head models bear a large degree of resemblance to the template and may not accurately capture fine anatomical features such as the exact shape of the gyri and sulci of the patient’s brain. The frequency of TTFields and the method of delivery (large transducer arrays) dictate that TTFields distribution is broad and coarse in geometry and distributes over the entire brain. Consequently, fine details in the anatomy are unlikely to affect the overall shape of the field distribution in the brain, and are therefore expected to have minimal effect on the analysis and conclusions presented in this study. This is in contrast to applications such as transcranial direct current stimulation (TDCS) [23], or deep brain stimulation [24], where it is important to accurately deliver the electric field to specific and often finely shaped anatomical structures.
The analysis performed in this study suggests a statistically significant connection between TTFields intensity at the tumor bed (multiplied by compliance) and patient outcome. This suggests that treatment planning in which array placement on the scalp is optimized in order to maximize TTFields intensity at the tumor bed could be important for improving patient outcome. Indeed, the NovoTAL system utilizes simple geometrical rules to optimize array placement. In the future, the algorithms presented in this chapter could form the basis for treatment planning software that uses realistic simulations of TTFields distribution in the body for treatment planning. TTFields treatment planning may even be extended beyond a calculation of positioning the arrays on the head. For example, planning may encompass surgical procedures, such as cranial remodeling, which have been shown to increase TTFields distribution at the tumor by over 50% in some cases [12].
In this study, we assumed homogeneous tissue properties for all tissue types based on average values reported in the literature, with the conductivity values assigned to the various compartments of the tumor. The assumption of homogeneity is reasonable when considering healthy tissue [see chapter on wEPT in this book]. However, glioblastoma tumors are structurally heterogeneous. Therefore, their electric properties are likely heterogeneous as well. However, very little literature investigating the electric properties of tumors exists. Such heterogeneity will affect TTFields distribution in the tumor bed. Future studies investigating the electric properties of tumors are planned so that tumor heterogeneity can be accounted for in simulations and treatment planning. Techniques such as wEPT [25] or MR-based electrical impedance tomography (MREIT) [26] provide spatial maps of the electric properties in tissue that could be extremely useful in this regard. However, their applicability to mapping electric properties in the 100 kHz-1 MHz frequency range should first be established. It is important to note that in the context of this study, the effect of heterogeneity in the tumor bed on TTFields dose, and its connection to outcome is effectively accounted for in the analysis through the averaging performed on field intensity in the tumor bed and the large number of patients in the study.
To conclude, this chapter presents novel methods for creating realistic head models suitable for the simulation of TTFields. The approach has enabled a ground-breaking study in which simulation and clinical data have been combined to establish a connection between TTFields dose at the tumor and patient outcome. Future studies based on our approach will lead to a better understanding of TTFields therapy, ultimately leading to improved patient outcomes.
References
Kirson, E. D., et al. (2004). Disruption of cancer cell replication by alternating electric fields. Cancer Research, 64(9), 3288–3295.
Kirson, E. D., et al. (2007). Alternating electric fields arrest cell proliferation in animal tumor models and human brain tumors. Proceedings of the National Academy of Sciences of the United States of America, 104(24), 10152–10157.
Giladi, M., et al. (2015). Mitotic spindle disruption by alternating electric fields leads to improper chromosome segregation and mitotic catastrophe in cancer cells. Scientific Reports, 5, 18046.
Stupp, R., et al. (2017). Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA, 318(23), 2306–2316.
Kanner, A. A., et al. (2014). Post Hoc analyses of intention-to-treat population in phase III comparison of NovoTTF-100A™ system versus best physician’s choice chemotherapy. Seminars in Oncology, Suppl 6, S25–S34.
Wenger, C., et al. (2015). The electric field distribution in the brain during TTFields therapy and its dependence on tissue dielectric properties and anatomy: A computational study. Physics in Medicine and Biology, 60(18), 7339.
Korshoej, A. R., et al. (2018). Importance of electrode position for the distribution of tumor treating fields (TTFields) in a human brain. Identification of effective layouts through systematic analysis of array positions for multiple tumor locations. PLoS One, 13(8), e0201957.
Chaudhry, A., et al. (2015). NovoTTFTM-100A system (tumor treating fields) transducer array layout planning for glioblastoma: A NovoTALTM system user study. World Journal of Surgical Oncology, 13, 316.10. FSL FLIRT.
Thielscher, A., et al. (2015). Field modeling for transcranial magnetic stimulation: A useful tool to understand the physiological effects of TMS? Milano: IEEE EMBS.
Korshoej, A. R., et al. (2016). Enhancing predicted efficacy of tumor treating fields therapy of glioblastoma using targeted surgical craniectomy: A computer modeling study. PLoS One, 11(10), e0164051.
Korshoej, A. R., Hansen, F. L., Thielscher, A., Von Oettingen, G. B., Christian, J., & Hedemann, S. (2017). Impact of tumor position, conductivity distribution and tissue homogeneity on the distribution of tumor treating fields in a human brain: A computer modeling study. PLoS One, 12(6), e0179214.
Timmons, J. J., et al. (2017). End-to-end workflow for finite element analysis of tumor treating fields in glioblastomas. Physics in Medicine and Biology, 62(21), 8264–8282.
Ashburner, J. (2012). SPM: A history. NeuroImage, 62, 791–800.
https://www.synopsys.com/simpleware/products/software/scanip.html
Wenger, C., et al. (2016). Improving tumor treating fields treatment efficacy in patients with glioblastoma using personalized array layouts. International Journal of Radiation Oncology, Biology, Physics, 94(5), 1137–1143.
Woo, J., et al. (2012). Reconstruction of high-resolution tongue volumes from MRI. IEEE Transactions on Biomedical Engineering, 59(12), 3511–3524.
Holmes, C. J., et al. (1998). Enhancement of MR images using registration for signal averaging. Journal of Computer Assisted Tomography, 22(2), 324–333.
Huang, Y., et al. (2015). Fully automated whole-head segmentation with improved smoothness and continuity, with theory reviewed. PLoS One, 10(5), e0125477.
Yushkevich, P. A., Piven, J., Hazlett, H. C., Smith, R. G., Ho, S., Gee, J. C., & Gerig, G. (2006). User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31(3), 1116–1128.
Ballo, M., Urman, N., Lavy-Shahaf, G., Grewal, J., Bomzon, Z., & Toms, S. (2019). Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: A large-scale numerical simulation-based analysis of data from the phase 3 EF-14 randomized trial. International Journal of Radiation Oncology, Biology Physics, In press.
Miranda, P. C., et al. (2017). Optimizing electric-field delivery for tdcs: Virtual humans help to design efficient, noninvasive brain and spinal cord electrical stimulation. IEEE Pulse, 8(4), 42–45.
Arle, J. E., et al. (2016). High-frequency stimulation of dorsal column axons: Potential underlying mechanism of paresthesia-free neuropathic pain relief. Neuromodulation, 19(4), 385–397.
Michel, E., Hernandez, D., & Lee, S. Y. (2017). Electrical conductivity and permittivity maps of brain tissues derived from water content based on T1 -weighted acquisition. Magnetic Resonance in Medicine, 77, 0094–1103.
Zhang, X., Liu, J., & He, B. (2014). Magnetic resonance based electrical properties tomography: A review. IEEE Reviews in Biomedical Engineering, 7, 87–96.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2019 The Author(s)
About this chapter
Cite this chapter
Urman, N. et al. (2019). Investigating the Connection Between Tumor-Treating Fields Distribution in the Brain and Glioblastoma Patient Outcomes. A Simulation-Based Study Utilizing a Novel Model Creation Technique. In: Makarov, S., Horner, M., Noetscher, G. (eds) Brain and Human Body Modeling. Springer, Cham. https://doi.org/10.1007/978-3-030-21293-3_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-21293-3_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21292-6
Online ISBN: 978-3-030-21293-3
eBook Packages: EngineeringEngineering (R0)