Abstract
MRI is considered the gold standard in soft tissue diagnostic of the lumbar spine. Number of protocols and modalities are used – from one hand 2D sagittal, 2D angulated axial, 2D consecutive axial and 3D image types; from the other hand different sequences and contrasts are used: T1w, T2w; fat suppression, water suppression etc. Images of different modalities are not always aligned. Resolutions and field of view also vary. SNR is also different for different MRI equipment. So the goal should be to create an algorithm that covers great variety of imaging techniques.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
MRI is considered the gold standard in soft tissue diagnostic of the lumbar spine. Number of protocols and modalities are used – from one hand 2D sagittal, 2D angulated axial, 2D consecutive axial and 3D image types; from the other hand different sequences and contrasts are used: T1w, T2w; fat suppression, water suppression etc. Images of different modalities are not always aligned. Resolutions and field of view also vary. SNR is also different for different MRI equipment. So the goal should be to create an algorithm that covers great variety of imaging techniques.
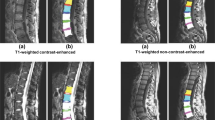
We consider the segmentation as the first step in a 3-step process: 1. Segmentation Fig. 1(a); 2. Measurements Fig. 1(b); 3. Diagnosis (in the case shown in the Fig. 1(b) - severity of disk herniation and central canal stenosis grading). Our system detects most of the visible tissues that are relevant for diagnosing a pathology. One of the tissues is Intervertebral Discs. We applied our method with some extensions to detect intervertebral discs in the IVDM3Seg Segmentation Challenge [7].
2 Methods
In order to cover different protocols a 2D single modality algorithm was developed that in cases of 3D multi-modality data can be used in ensemble of multiple 2D single modality data models. Our 2D algorithms are using CNN [1] and are greatly inspired of ResNet [2]. Dropout regularization [3] and batch normalizations [4] are also used. FCN [5] style network is used for the super-pixel classification as this enable arbitrary field of view input. Super-pixels are 8 times smaller (in all dimensions) than the actual pixels (as stride 8 is used due to 3 2 × 2 max pooling layers). As a result fine grained details are lost, so we use Unet-like [6] architecture for up-scaling the low resolution map into the resolution of the input image. Separate 2D single modality results are combined into 3D results using ensemble with learnable weights combining the 3D information from 3 separate probability maps. To overcome the problem with small data set size, extensive augmentations were used: crop and resize, tilt, rotate, dynamic range changes, random noise in all possible combinations.
Datasets.
Our dataset consists of 30 patient studies, 918 axial and sagittal slices in total. The patients’ age ranges from 30 to 50 years old, with a mean age of 37.5 years old, including both male and female patients suffering from lower back pain. This data was provided by three medical centers, two of which use a GE Medical Systems to acquire MRI. The third set of MRI was acquired by a SIEMENS machine. The characteristics of the slices in the dataset vary:
-
Voxel Thickness: 3.5 mm to 10 mm (mean 7.4 mm)
-
Repetition Time: from 1040 ms to 6739 ms
-
Echo Time: from 9.6 ms to 110.3 ms
-
Axial Resolutions (Cols × Rows): 512 × 512, 276 × 192
-
Sagittal Resolutions (Cols × Rows): 512 × 512, 384 × 768.
The slices are not uniformly spaced and are not parallel to each other. The axial slices are parallel to each of the discs. This way there is no value for each of the voxels in the volume of the study. 3D ensemble from this data set is not straight forward The challenge dataset provided by IVDM3seg consisting of 16 patients with full 3D data available, consisting of 4 modalities.
Feature Extraction.
Fully convolutional ResNet-50 was used for feature map extractor. The model is pretrained on COCO. The minimal stride of the feature map for pretrained model we could find was 8. For Image (512 × 512 × 3) a feature map (64 × 64 × 1024) is produced. It is believed that the bigger feature stride causes lower resolution imperfections in the masks. For 256 * 256 * 37 the smallest feature map resolution is 4. An attempt to overcome this limitation was UNet-like mask predictor architecture.
The model originally uses 3 input channels (RGB). To make use of the modalities of the MRI, the 3 most useful modalities are used as input in the feature extractor model (Fig. 2).
Segmentation.
To produce more accurate masks, which capture finer detail and higher frequency changes in the contour, a UNet-like architecture was used. To produce the mask, series of up-convolutions are used, starting from the stride 8 feature maps of the feature extractor and doubling the resolution on each layer. Each layer is combined with the corresponding resolution feature map from an internal layer in the feature extractor. This way higher level, lower resolution semantic features are used as context, and lower level, higher resolution features are used for finer details (Fig. 3).
3D Ensemble.
The prediction from 3 2D models (axial, sagittal, coronal) are combined using a 3D convolution neural network. It has 2 layers of 3 × 3 × 3 convolutions that are trained on the predictions of one part of the validation set and validated on the other part. The challenge data set is divided into train and validation set. The 3 models are trained on the train set and the hyper parameters are tuned on the validation set. When the models are trained one part of the validation set is predicted and the predicted probability maps are used to train the 3D convolution model.
The input for the 3D convolution is 6 channel 3D matrix. Each plane has 2 channels. Probability of segmentation of disk and some relative position encoding parameter. The position encoding parameter helps the model combine the information from all the 3 models in the best way.
The 3D ensemble combines the best predictions form each single plane 2D model. Each model is better than the others at some specific regions of the disc and worse in others. A 2D model mask is better at the middle section (according to the direction of the normal of the plane) of the disc than in the endings (where the intersections are smaller). By putting more weight on the proper model (plane) prediction at each region, the 3D ensemble mask combines the best prediction from each of the models in each region. So the combined mask is better than any single plane 2D mask.
The other big effect of the 3D model is that it filters some prediction noise. 2D models sometimes predict false positives. There is a low probability that in a particular voxel more than 1 models have predicted false positives so the noise gets filtered. Single voxel or some small objects gets filtered too (Fig. 4).
Augmentation.
In order to train a big model with a small dataset, extensive augmentations were used. Elastic whole image deformation – Take N × N uniform grid of points on the image, and chose a random direction vector for each point. Move each pixel in that direction with amplitude, proportional to f(inverse distance to the point). Tissue deformation – chose random points on the contour of the object and inside the object. Apply the elastic deformation on that points.
All the above augmentations, augment the image as well as the GT mask. Tissue brightness – change the values of the pixels lying in the ground-truth mask in some random direction. Noise – add white noise to each image, without changing the GT masks (Fig. 5).
3 Results
Challenge Result.
Results achieved by 4-fold cross validation using the data provided by the organizers [7] are listed in Table 1.
As seen in Table 1, the 3D ensemble outputs 2 times less errors in the mask.
It is interesting that the middle discs have bigger dice than the first and the last. This phenomenon is observed in the other participants in the challenge too. May be the middle disc are “easier”. We had big problems with detecting the 7-th disc (Th11–Th12) on some patients. Because only 7 discs were labeled in the GT, in some patients unlabeled discs appeared above the 7-th disc, which caused our classifier to get confused if it has to detect discs closer to the top end of the image or not. The problem was overcome by cropping the unlabeled discs out of the GT images during training. Test time is 3:10 s on a single GPU machine that can be reduced to 1:20 in batch mode. The model supports resolution of 512/512 that is 4 (2 × 2) times bigger than necessary for this particular set so test time can be further reduced (Table 2).
During the cross validation we never observed detection of the sacrum. With this assumption we developed a simple filtration algorithm which takes the bottom 7 discs. But in test set evaluation the sacrum was detected in two of the patients which, led to missing the top disc completely and punishing the metrics of the bottom disc (Table 3).
We calculated our expected test set dice metric without detecting the sacrum and missing the top disc (Table 4).
Results in detecting disc herniation are as follows (Tables 5, 6 and 7).
The performance of disc segmentation on our data is slightly worse than in the challenge because of the acquisition format that we use. The data in the challenge is 3D and has info for every voxel, while our data has slices spaced on bigger distance. The axial slices and sagittal slices are different sequences which are not strictly orthogonal to each other and to the coordinate system axes. 3D ensemble of the axial and sagittal slices is not straight forward. An experiment was undertaken to find the relation between the training set size and the validation set performance (Fig. 6).
4 Discussion
It seems like the ground truth masks of the 3D data are labeled only in the sagittal plane. The human annotator labeled each sagittal slice of the 3D matrix, producing 3D ground truth matrix concatenated from all the sagittal slices. This leads to strange artifacts when looking the 3D matrix form other perspectives.
The same thing is possible during prediction. We have 3 2D models each working on one of the three planes. Each model can make 1–2 pixel mistakes in the edge. So the 3D convolution, that combines the outputs of the three models, uses the prediction of each of the model in the region where it is most accurate. This way using the strengths of each of the model (Figs. 7 and 8).
The same thing is possible during prediction. We have 3 2D models each working on one of the three planes. Each model can make 1–2 pixel mistakes in the edge. So the 3D convolution, that combines the outputs of the three models, uses the prediction of each of the model in the region where it is most accurate. This way using the strengths of each of the models.
Problems with Detection.
Although the mask quality of correctly detected objects is relatively good, there is a problem with detecting small objects. The most likely reason is the big feature map stride (8 × 8). When the stride is big, one feature map pixel corresponds to bigger area of original pixels. Instead of training one convolutional filter many times, one filter gets trained less times, but with more diverse set of positions of the smaller object in it. So there are a lot of places where the object did not appear in the filter’s field of view. This leads to underfitting of the bigger convolutional filter and to underfitting of the detector.
5 Conclusion
Test accuracy was similar to the previously reported results using 3D convolutions on the test data of the previous challenge [8] although the algorithm was designed for 2D single-modality data. Training set accuracy is near 100% which can be expected as the complexity of the model is very big and definitely high variance is the current drawback. Never the less we decided to not reduce the complexity as we believe bigger training set is necessary for reaching human level accuracy. So further test set accuracy improvements can be expected by increasing the training set. Our intention for the future development is to cover great variety of tissues and pathologies by acquiring an annotated training set of 500 patients. Part of them will be released to the scientific community.
References
Le Cun, Y., Bottou, L., Bengio, Y.: Reading checks with multilayer graph transformer networks. In: ICASSP 1997, vol. 1, pp. 151–154. IEEE (1997)
He, K., et al.: Deep residual learning for image recognition. In: CVPR 2016 (2016)
Hinton, G.E., et al.: Improving neural networks by preventing co-adaptation of feature detectors. Technical report. arXiv:1207.0580
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. CoRR abs/1502.03167 (2015)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. arXiv:1411.4038 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Chen, C., Belavy, D., Zheng, G.: 3D intervertebral disc localization and segmentation from MR images by data-driven regression and classification. In: Wu, G., Zhang, D., Zhou, L. (eds.) MLMI 2014. LNCS, vol. 8679, pp. 50–58. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10581-9_7
Xiaomeng, L., et al.: 3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR images. Med. Image Anal. 45, 41–54 (2018)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Georgiev, N., Asenov, A. (2019). Automatic Segmentation of Lumbar Spine MRI Using Ensemble of 2D Algorithms. In: Zheng, G., Belavy, D., Cai, Y., Li, S. (eds) Computational Methods and Clinical Applications for Spine Imaging. CSI 2018. Lecture Notes in Computer Science(), vol 11397. Springer, Cham. https://doi.org/10.1007/978-3-030-13736-6_13
Download citation
DOI: https://doi.org/10.1007/978-3-030-13736-6_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-13735-9
Online ISBN: 978-3-030-13736-6
eBook Packages: Computer ScienceComputer Science (R0)