Abstract
The development of antimalarial drugs requires performing laboratory experiments that include the analysis of blood smears infected with Plasmodium berghei. Analyzing visually the resulting microscopy images is usually a slow and tedious task prone to errors due to fatigue and subjectivity of the analysts. These facts motivated the creation of digital image processing systems to automate the aforementioned analysis. We present in this work a computer vision solution which processes microscopy images of blood smears. This system performs tasks like illumination correction, color compensation, image segmentation including separation of clumped objects and the extraction and selection of color features. Then a set of classifiers was tested to find the best one in terms of classification results. Here a new feature named pixels fraction was introduced and a number of other color features were extracted, from which a subset was selected for the classification of the cells into either normal or infected. The classifiers tested for this application were: support vector machines (SVM), K-nearest neighbors (KNN), J48, Random Forest (RF), Naïve Bayes and linear discriminant analysis (LDA). All of them were evaluated in terms of their performance expressed as correct classification rate, sensitivity, specificity, F-measure and area under Receiver Operating Characteristic (ROC) curve (AUC). The usefulness of the pixels fraction as a new and effective feature was demonstrated by the experimental results. In regard of classifiers, J48 and Random Forest showed the best results.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
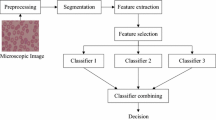
Malaria is an infectious disease showing high degrees of morbidity and mortality, for which the World Health Organization estimated 215 million of infected persons and 445000 deaths in 2016 [1]. This serious health problem claims for new diagnose tools and anti-malarial drugs. Microscope analysis of large amounts of blood smears in order to detect the presence of the Plasmodium parasite is a problem of primary importance both to diagnose the disease in humans and to determine the infection rate in laboratory mice during the process of developing anti-malarial drugs. This analysis, when made by human experts is a slow and tedious process whose results are prone to errors due to tiredness, subjectivity and to the probable low rate of positive cases (infected erythrocytes). This has motivated developing digital image processing (DIP) - computer vision (CV) solutions for this process, which is the topic addressed in this work.
There are various published works on this problem, usually implementing diverse image processing procedures to obtain appropriate image features and afterwards performing the classification of the erythrocytes, examples of which can be found in [2,3,4,5]. These procedures include tasks such as image conditioning through non-uniform illumination correction, filtering and color normalization. Image segmentation of the microscope digital images of blood smears is essential to separate erythrocytes from other blood components and artifacts, as well as to appropriately separate clumped (touching and overlapping) erythrocytes. After this, there have been different approaches to obtain appropriate features from segmented erythrocytes, to ensure an effective classification. Finally, testing and selecting effective classifying algorithms complete the design of the system. Examples of this can be found in [6,7,8,9]. Classifiers like linear discriminant analysis (LDA), K-nearest neighbors (KNN), support vector machines (SVM) and others have been used for this purpose.
The contribution of this paper consists in finding new color features with high discriminating capabilities, combined with a study of their best possible combination with appropriate classifier algorithms. The system is oriented towards applications to anti-malarial drug development, in which the analysis of blood smears from laboratory mice demands a low rate of false positives.
2 Materials and Methods
2.1 Sample Images
The images used in this research were taken from Giemsa-stained blood smear slides from mice experimentally infected with Plasmodium berghei, kindly donated by Dr. José Antonio Escario García Trevijano from the Faculty of Pharmacy, Universidad Complutense de Madrid. A Zuzi 122/148 tri-ocular microscope was used, equipped with a Microscopy 319 CU digital camera with 3.2 MP resolution and 8-bit RGB output without compression, producing a 2048 × 1536 pixels matrix, with pixel size 3.2 × 3.2 μm, signal to noise ratio 43 dB and optical magnification 50×. The digital images were saved in .tiff (tagged image file) format. An annotated database was created with the aid of two expert analysts from CBQ. This database is intended to perform all the DIP-CV procedures to obtain the features, training the classifiers and realizing tests to assess the effectiveness of classification. A total of 211 images were obtained, from which a set of 600 images of independent segmented erythrocytes was formed, comprising 400 un-infected and 200 Plasmodium-infected cells, for which examples are shown in Fig. 1. Notice the reddish-purple spots inside individual erythrocytes that harbor the parasites.
The size of the sample set was determined following [10] as a minimum necessary to obtain a reasonable error when evaluating the correct classification rate CCR, when it is expected to be above 0.95 for the classifiers evaluated. These numbers attempted also to cope with a possible class-imbalance. The image sizes of individual cells depend on their physical size, which can exhibit certain natural variability and can also be affected by the presence or not of the parasite.
2.2 Image Conditioning
All the images were initially acquired in the RGB color space with 8 bpp/channel and the intensity of the color components was normalized to the interval [0, 1]. They were converted afterwards to the HSI color space. Other pre-processing steps applied were [3 × 3] median filtering to the intensity component and a morphological top hat with an appropriate structuring element to compensate any possible illumination imbalance. Conversion of the images to the La*b* space was made also after segmentation to allow obtaining more features. Information on color spaces is given in [11].
2.3 Segmentation
Segmentation of erythrocytes was performed in two steps. Firstly, the Otsu’s algorithm as used in [2] was applied in this case adaptively to intensity component of the image by dividing it into 16 patches that were segmented independently. This coarse segmentation binarized the image into foreground objects (cells, including clumps) and background. Then the cell clumps were separated (fine segmentation) employing a modification of the algorithm described in [12], using weighted outer distance and marker-controlled watershed transforms, with the regional maxima of the distance transform as internal markers. This process proved to be effective in accurately detecting and splitting the cell clumps. Other components of the blood smears like leukocytes and platelets were eliminated using the procedure described in [6] and other artifacts were suppressed as well by morphological area opening, using a threshold derived from the median size of the erythrocytes.
2.4 Color Normalization
Color-based features obtained from the images are essential here for the classification process. In microscopy images, color can be altered due to changes in the illumination source and to the procedure of preparing the samples. This led to the necessity of color normalization by means of DIP techniques. Here the method described in [4] was used for this purpose and the results are illustrated in Fig. 2.
2.5 Feature Extraction and Pixels Fraction
As we stated previously, the classification process was performed here on the basis of color features solely. A total of 13 features were obtained for each of the RGB, HSI and La*b* color spaces. These were, for each color component: mean, variance and skewness as described in [13], as well as kurtosis and a new feature whose introduction is the main contribution of this work: the pixels fraction. Considering the three color spaces this led to a total of 39 features.
The pixels fraction is defined here based in the relative coincidence of the pixel values of the target and a reference, in the planes corresponding to the three color channels, for a specific color space. A small set of regions of interest (ROI) located inside the reddish-purple colored region characteristic of the parasites in a color-normalized erythrocyte were taken as a reference, as shown in Fig. 3. For this set of regions, the mean μc and standard deviation σc of the intensity in each color channel are determined, where C can take the values R (red), G (green) or B (blue). To illustrate the calculation of the pixels fraction in the RGB color space, consider a cell being analyzed. Then the number of pixels is determined for it, whose intensities \( imC \) corresponding to the three color components satisfy simultaneously the condition
In Eq. 1, the factor 2 multiplying \( \sigma_{c} \) widens the acceptance intervals for the color components of a given pixel and was determined heuristically. The pixels fraction \( p_{f} \) is finally determined for the image of an erythrocyte in a specific color space by dividing the number of pixels nf satisfying the condition 1 by the total number N of pixels in the image.
The value of \( p_{f} \) was determined analogously in the HSI and La*b* color spaces.
2.6 Feature Selection and Classification
In this step Weka 3.9 [14] facilities were used. Firstly, a selection from the erythrocyte features previously described, by means of filtering (CfsSubsetEval with a greedy stepwise search method) was used. This selected seven features. Then, a ranking alternative (InfoGainAttributeEva) allowed using the first 20 ranked features as well as the first 7 (to match the number selected through filtering) for classification, as well as all the features. Then the effectiveness of these alternatives were compared.
Classification was then made comparing the following algorithms: SVM, KNN, J48, Random Forest (RF), Naïve Bayes (NB) and Linear Discriminant Analysis (LDA). In the case of SVM and KNN various alternatives in their parameters (polynomial and PUK kernels in SVM, K = 1, 3, 5, 7 for KNN) were tested and those with the best results were used in the comparison to the rest of the classifiers.
The comparison among the various classifiers was performed by ten-fold cross-validation and 1/3−2/3% split. The indexes of effectiveness used were the correct classification rate (CCR), sensitivity (Se), specificity (Sp), F-measure and AUC. Finally a more realistic experiment was performed considering the possibility of defining visually dubious cases as a third class, a situation often encountered in practice due to spurious colored pixels. In this case the results were expressed in terms of confusion matrices. All the features were previously normalized.
3 Results and Discussion
All the steps described in Sect. 2 were performed for the dataset composed of 600 erythrocytes that was mentioned earlier. Special attention was paid to building the (600 × 39) feature matrix of this dataset.
3.1 Feature Selection
Results of feature selection by using the two methods (filter and ranking) are shown in Table 1. Notice that despite in general the seven first features ranked by the InfoGainAttributeEva method differ from those selected by CfsSubsetEval, in both cases the pixels fraction in the three color spaces used were the first ones in the list, which confirms their usefulness.
3.2 Classification
Classification results by using 7 features obtained through ranking and filtering, are shown in Tables 2 and 3, respectively. In this case the performance measures used in a 10-fold cross-validation experiment were CCR, Se, Sp, F-measure and AUC. Classification results using the whole set of features or the first 20 in the ranking list, not shown due to space limitations, were inferior to those shown in the tables. This suggests that there is some degree of noisy behavior in the discarded features whose deletion improved the classification results. Some variants of SVM and KNN were disregarded previously in favor of those included in the tables, which exhibited better behavior. Notice that the best performance was obtained by the J48 and Random Forest classifiers, which yielded results close to 100%.
Notice that the pixels fraction should be theoretically zero for a normal erythrocyte. This could lead to the idea that classification of a cell is a trivial task. However, in practice some spurious colored pixels could appear and provoke an erroneous classification. This motivated here to employ a larger set of color-based features that could provide classification improvements in these cases, as well as introducing a third “dubious” class. Table 4 shows the confusion matrix obtained in the classification process when considering this third class. This is important because, differently to malaria diagnose in humans, when determining the infection rate in laboratory mice through microscopy analysis, which is the target of this work, dubious cells are usually disregarded by human analysts. Following the same procedure as before, in this case only four features (pixels fraction among them) were chosen by the filter selector. When using the J48 and RF classifiers, almost all dubious cases were correctly classified, all normal cells were still classified as normal and a small proportion of infected erythrocytes were classified as dubious.
4 Conclusion
Automated classification of erythrocytes to detect the presence of Plasmodium berghei parasites is a very important task in anti-malarial drug development. This is currently an open area of research and this work presents two contributions in this area. The first one has been an improvement of the use of color information in the classification process by means of the definition of a new feature, called the pixels fraction, whose effectiveness was proved by two facts. Firstly, its values for the three color spaces involved in this study (RGB, HSI and La*b*) were selected among the most important features by both the filter and the ranker feature selectors used. Secondly, the classification results using the pixels fraction were remarkable. Several classifier algorithms were tested among which J48 and RF exhibited the best results in terms of the evaluated measures of performance. The second contribution was linking a set of image processing steps with the classifiers, to complete a computationally efficient way to classify erythrocytes in malaria studies. Future work will address an evaluation of the effectiveness of Convolutional Neural Networks classifiers for the application studied in this work.
References
World Health Organization. World Malaria Report (2017)
Arco, J.E., Górriz, J.M., Ramírez, J., et al.: Digital image analysis for automatic enumeration of malaria parasites using morphological operations. Expert Syst. Appl. 42, 3041–3047 (2015). https://doi.org/10.1016/j.eswa.2014.11.037
Abdul-Nasir, A.S., Mashor, M.Y., Mohamed, Z.: Colour image segmentation approach for detection of malaria parasites using various colour models and k-means clustering. WSEAS Trans. Biol. Biomed. 10(1), 41–55 (2013)
Tek, F.B., Dempster, A.G., Kale, I.: Parasite detection and identification for automated thin blood film malaria diagnosis. Comput. Vis. Image Underst. 114, 21–32 (2010). https://doi.org/10.1016/j.cviu.2009.08.003
Das, D.K., Maiti, A.K., Chakraborty, C.: Automated system for characterization and classification of malaria-infected stages using light microscopic images of thin blood smears. J. Microsc. 257, 238–252 (2015). https://doi.org/10.1111/jmi.12206
Di Ruberto, C., Dempster, A., Khan, S., Jarra, B.: Analysis of infected blood cell images using morphological operators. Image Vis. Comput. 20, 133–146 (2002). https://doi.org/10.1016/S0262-8856(01)00092-0
Ajala, F.: Comparative analysis of different types of malaria diseases using first order features. Int. J. Appl. 8, 20–26 (2015). https://doi.org/10.5120/ijais15-451297
Loddo, A., Di Ruberto, C., Kocher, M.: Recent advances of malaria parasites detection systems based on mathematical morphology. Sensors 18(2), 513 (2018). https://doi.org/10.3390/s18020513
Chavan, S., Nagmode, M.: Malaria disease identification and analysis using image processing. Int. J. Latest Trends Eng. Technol. 3(3), 218–223 (2014)
Walpole, R.E., Myers, R.H., Myers, S.L., Keying, E.Y.: Probability and Statistics for Engineers and Scientists: Pearson New International Edition. Pearson Higher Education, Upper Saddle River (2013)
Gonzalez, R.C., Woods, R.E.: Digital Image Processing, 3rd edn. Pearson Prentice Hall, Upper Saddle River (2008)
Jierong, C., Rajapakse, J.C.: Segmentation of clustered nuclei with shape markers and marking function. IEEE Trans. Biomed. Eng. 56(3), 741–748 (2009). https://doi.org/10.1109/TBME.2008.2008635
Saikrishna, T.V., Yesubabu, A., Anandarao, A., Rani, T.S.: A novel image retrieval method using segmentation and color moments. Adv. Comput. 3(1), 75–80 (2012). https://doi.org/10.5121/acij.2012.3106
Bouckaert, R., Frank, E., Hall, M., et al.: WEKA Manual for Version 3-6-13. CreateSpace Independent Publishing Platform (2015)
Acknowledgment
The authors acknowledge the VLIR-UOS Project Cuba ICT Network for the financial support provided to this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Lorenzo-Ginori, J.V. et al. (2019). Color Features Extraction and Classification of Digital Images of Erythrocytes Infected by Plasmodium berghei. In: Vera-Rodriguez, R., Fierrez, J., Morales, A. (eds) Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. CIARP 2018. Lecture Notes in Computer Science(), vol 11401. Springer, Cham. https://doi.org/10.1007/978-3-030-13469-3_83
Download citation
DOI: https://doi.org/10.1007/978-3-030-13469-3_83
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-13468-6
Online ISBN: 978-3-030-13469-3
eBook Packages: Computer ScienceComputer Science (R0)