Abstract
We propose a deep learning-based pipeline that, given a low-dose computed tomography of a patient chest, recommends if a patient should be submitted to further lung cancer assessment. The algorithm is composed of a nodule detection block that uses the object detection framework YOLOv2, followed by a U-Net based segmentation. The found structures of interest are then characterized in terms of diameter and texture to produce a final referral recommendation according to the National Lung Screen Trial (NLST) criteria. Our method is trained using the public LUNA16 and LIDC-IDRI datasets and tested on an independent dataset composed of 500 scans from the Kaggle DSB 2017 challenge. The proposed system achieves a patient-wise recall of 89% while providing an explanation to the referral decision and thus may serve as a second opinion tool to speed-up and improve lung cancer screening.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Lung cancer is the deadliest type of cancer worldwide in both men and women [1] but early diagnosis significantly increases patient survival rate. In fact, the National Lung Screen Trial (NLST) showed that annual screening of lung cancer risk groups with low-dose chest computed tomography (LDCT) via manual analysis of scans by experts reduces lung cancer mortality by 20% [2]. However, LDCT screening is challenging because (i) the process is prone to errors due to factors such as interobserver variability and (ii) the equipment and personnel costs of these programs inhibit their application on developing countries, where tobacco consumption is difficult to control [3]. To address the problem of lung cancer detection we propose a second opinion system that helps to reduce the overall screening burden by indicating if a patient should or not be referred for follow-up. The framework is composed of an initial nodule detection step, nodule segmentation and finally a scan-wise decision based on the NLST criteria for positive screens and thus provides an interpretable justification of its decision.
Lung nodule detection is a mandatory step for the automatic referral pipeline. Deep learning has become the standard technique to complete this task, with the leaderboard of LUNA16Footnote 1 nodule detection challenge being mainly composed of these kind of approaches [4]. The most common pipeline is to detect candidates by slice or 3D-wise via an object detection framework such as Faster-R CNN [5] followed by a 3D convolutional neural network (CNN) for false-positive (FP) reduction and these systems achieve detection sensitivities greater than 0.80 with 0.125 FP/scan or greater than 0.90 with 1 FP/scan. Then, nodule segmentation is used for characterizing the detected abnormalities. The most common approach for nodule segmentation is to use intensity (or HU) and shape features [6], but CNNs can also be applied for 3D nodule segmentation and achieve Sorensen-Dice coefficients close to 0.8 [7].
Despite the advances on both nodule detection and segmentation, automatic patient referral is little explored. Recently, KaggleFootnote 2 hosted a challenge aimed at the development of algorithms for predicting if a patient should be referred for follow-up after screening. The training set is composed of 1398 training scans with labels at the scan level. It is widely known that the majority of the scans originated from the NLST trial, but the exact origin of each anonymized scan has not been disclosed by Kaggle. In this challenge, the best methods used deep learning approaches with an initial candidate detection followed by an expected malignancy prediction that allow to achieve an overall log loss of 0.39–0.41.
We move beyond the nodule detection task and aim at tackling the more complex lung cancer screening pipeline. Our contribution to the state-of-the-art is a single step nodule detection algorithm followed by a segmentation and field-knowledge classification step that allows a near-human scan-wise abnormality detection performance for scan referral. The next section describes the developed algorithms and the datasets used for validation. Section 3 discusses the performance of the proposed system. Finally, Sect. 4 summarizes our study.
2 Method
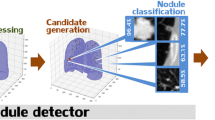
Our system was designed to follow the standard clinician pipeline of NLST [2], where radiologists were instructed to refer a patient for follow-up if any non-calcified nodule with diameter \(d>4\) mm was found. With that in mind, our system is composed of 3 main steps (see Fig. 1) (i) nodule detection via YOLOv2 (Sect. 2.2) that focuses on nodules with \(d>4\) mm, (ii) nodule segmentation for measurement and characterization via U-Net (Sect. 2.3) and (iii) scan-wise referral indication based on the NLST guidelines (Sect. 2.4).
2.1 Datasets and Technical Details
The nodule detection network is trained on the LUNA16 dataset [4], which contains 888 scans from the LIDC-IDRI dataset [8]. The LIDC-IDRI contains 1012 LDCT scans with variable slice thickness and nodule voxel-wise annotations from up to 4 different expert radiologists. The LUNA16 contains the information of 1186 nodules’ centroids and diameter (no voxel-wise data) with an agreement level 3 or higher, as well as the centroid of non-nodule lesions of diagnostic interest. For the nodule detection step, we train in subsets 1–9 (20% validation) and test on the 89 scans of subset 0. The test set has 112 nodules, from which 80 have \(d>4\) mm. The nodule segmentation system is trained on LIDC-IDRI with an agreement level 1 or higher and multiple segmentations are combined via logical OR. We train on 1400 axial view \(64\times 64\) pixels (approx. \(51\times 51\) mm) patches, validate on 300 and test on 570 samples. We experimentally set the segmentation threshold at 0.5 by analysis of the results on the validation set.
We tested our screening system on 500 randomly selected scans from the \(1^{st}\) stage of the Kaggle dataset. The scans are labeled according to future cancer presence (123 cases) or low cancer risk (377 cases). All datasets are anonymized and there is no access to relevant patient metadata. Unless stated otherwise, we consider the HU interval [\(-1000\), 400] for our experiments.
Experiments were performed on a Intel Core i7-5960X @3.00 GHz, 32 Gb RAM, \(2\,\times \) GTX1080 desktop. The framework was developed on Python 3.5 and Keras 2.0.4. The YOLOv2 implementation is based onFootnote 3. Both YOLOv2 and U-Net were trained with optimizer Adam (learning rate 1e–4) and we used real time data augmentation by randomly applying translations, zooms, edge sharpening, blurring and brightness and contrast alterations to the training data.
2.2 YOLOv2 for Lung Nodule Detection
Our framework uses YOLOv2 [9], an end-to-end 2D object detection network, to perform lung nodule detection without the need for a FP reduction step (refer to Fig. 2 for an example). Specifically, the network divides the input image in a grid and predicts, for each grid element, how likely there is a nodule there. Let our input image be divided in blocks of size defined by the network architecture. Each block can contain a nodule, which bounding box we impose to be \(35\times 35\) mm (\(51\times 51\) pixels) to fit the largest nodules in the dataset. The model works as follows: (1) the InceptionV3 [10] network is used for extracting a \(M\times M\times N\) feature map, F, where M is the spatial grid size and N is the number of feature maps from the input image; (2) F is convolved into a \(M\times M\times 7\) new tensor, Y. Each element \(m_{i,j,k}\in Y_k\) has a direct correspondence with a block from the input image and each of the 7 feature maps corresponds to a variable of the respective \(\text {bounding box}_{i,j}\) to be optimized. The maps are responsible for controlling the probability of the box belonging to the ‘nodule’ or ‘other’ (non-nodule) classes (\(p_{i,j}(\text {nodule})=\sigma (m_{i,j,1})\)), how likely there is a nodule on that block (\(\text {confidence}_{i,j}=\sigma (m_{i,j,3})\)), the box width/height (\(\hat{w}_{i,j}=we^{m_{i,j,4}}\), \(\hat{h}_{i,j}=he^{m_{i,j,5}}\)) and the box center (\(\hat{x}_{i,j}=x_{i,j}+\sigma (m_{i,j,6})\), \(\hat{y}_{i,j}=y_{i,j}+\sigma (m_{i,j,7})\)), where \(\sigma \) is a sigmoid function. The entire network can be trained end-to-end by minimizing the following loss function:
where \(\mathcal {L}_1\) is the squared error and \(\mathcal {L}_2\) is the log loss function. In the end, only the boxes with high confidence and class probability are kept, as depicted in Fig. 2.
Since Inception V3 is pre-trained with RGB images, we train our model with images of the axial slice containing the nodule centroid together with one slice above and one slice below (2.5D). This strategy provides extra context to the network and has already been successful for lung nodule detection [11]. However, preliminary experiments led us to conclude that the selection of the non-central slices greatly hinders the nodule detection performance of YOLOv2. Namely, since all datasets show variable inter-scan slice thickness, the usage of the two immediate adjacent slices may provide a poor and irregular depth information. Instead, we retrieve slices from an approximate distance of 2 mm. This improved the system’s performance by almost 20% since it allows blood vessels, natural nodules’ confounders in the axial plane, to be better distinguished from the structures of interest (see Fig. 3) without compromising the model’s speed.
Influence of slice depth for the reduction of confounders in 2.5D. Red: top slice; Green: middle slice; Blue: bottom slice Left: original slice thickness (approx. 1 mm); Right: with approximately 2 mm slice thickness both small and large blood vessels are more visible in the red and blue channels. (Color figure online)
Training Details. Having in account the goal of our system, we opt for a very low value of FP/scan, close to 0.25, and thus higher risk of nodule detection failure because (i) detecting one nodule per scan (assumed to be lung cancer representative) is enough for referral and (ii) a large number of FP can lead to non-proper scan referral and thus unnecessary medical procedures. Based on these assumptions, we adopt a training scheme that achieves high scan-wise referral recall and specificity rates with a low number of FP detections.
First, we train the detection module using \(512\times 512\) 2.5D axial images that contain \(d>3\) mm nodules and use it for finding nodule candidates on all 2.5D axial images of the training scans. The resulting 2D predictions are combined to 3D nodule locations having in account the intra and inter-axial slice distance. Specifically, candidates are merged if there is more than 80% area overlap in the 2 adjacent slices. These 3D candidates integrate a second dataset composed of (1) all nodules from the training data with diameter \(\ge 4\) mm and (2) a set of nodules’ confounders composed of FP predictions with probability higher than 0.5 and all nodules with \(d<4\) mm. YOLOv2 is then retrained from scratch so that the weights can adapt to the two class problem, thus significantly reducing the final number of FP detections. This framework avoids the need for a second classifier for FP reduction and thus extra training-related parameter tuning. Similarly to the training step, scan-wise predictions are made by merging depth-wise the candidates that resulted from sliding the model over the scan.
2.3 U-Net for Nodule Segmentation
We segment the found nodules via an adaptation of U-Net [12]. Our model has 5 contracting steps, a \(1\times 1\) bottle neck and a higher number of feature maps on the expansive part. Also, Batch Normalization is performed at each convolutional layer for regularization. We use the soft intersection over union (IoU) as loss
where S and \(\hat{S}\) are the ground truth and the segmentation prediction \(\in [0,1]\).
2.4 Rule-Based Classification
Finally, we perform a referral decision based on the guidelines of NLST. Specifically, a decision tree is used for indicating if a patient should be referred for further examination or not. First, scans where no nodule candidates are detected are considered as negatives. From the remaining, a scan is considered pathological if there is at least a nodule candidate with \(d>4\) mm and less than 50% of calcified area. The nodule diameter d is the equivalent diameter of our segmentation, thus the diameter of a perfect sphere with an equal volume as the volume of our segmentation. The calcified area is computed by calculating the total area of all volumes above a threshold of 70 HU.
3 Experimental Results and Discussion
Our pipeline is composed of a novel single-step nodule detection system, followed by U-Net for segmentation and a final referral decision based on the NLST guidelines. Table 1 compares the performance of our system with the the top-3 methods from LUNA16. A nodule is considered detected (TP) if the distance to the prediction centroid is less than the nodule radius and hits on non-nodule lesions are not considered as FP. Also, note that we only consider nodules of \(d>4\) mm, instead of \(d>3\) mm, since these are the ones with relevance for screening. Even though it is not possible to state that our system is as good as other approaches because the detection-per-radius performance is not publicly available, the achieved recall is satisfactory for a 2.5D single-step nodule detection framework that requires less model-related parameter tuning and computation power than other state-of-the-art methods. Furthermore, our nodule detection achieves a scan-wise recall (i.e., finding at least one of the nodules in a scan) of 0.90, increasing to 0.95 if only nodules of \(d>4\) mm are considered.
U-Net achieves an average test IoU of \(0.63\pm 0.02\) and Sorensen-Dice coefficient of \(0.79\pm 0.15\), which is line with the state-of-the-art performance. Moreover, the estimated diameter error is of \(1.89\pm 3.20\) mm, with greater errors occurring at the largest nodules. This means that the system is capable of providing a robust measurement that does not compromise the NLST rule-based decision.
Figure 4 shows examples of nodules and the respective segmentation predicted by our method on the independent NLST dataset. As depicted in Fig. 4A–B, we are capable of detecting nodules of different sizes and challenging textures and still provide a good segmentation. Furthermore, in Fig. 4B we show examples of calcified structures being correctly detected due to the field knowledge-based threshold on the soft tissue HU window.
In terms of referral, we evaluate our system in terms of scan sensitivity and specificity to ease the comparison with the specialists from NLST. We do not compare with Kaggle solutions because our method does not produce a referral probability and thus log loss computation is not possible. Our screening pipeline achieves a cancer detection sensitivity of 89.4%, which is in line with the findings of the NLST study. This suggests that the proposed system is successfully locating, measuring and applying the decision criteria to relevant abnormalities as depicted in Fig. 4A–B. For comparison purposes, NLST specialists were capable of successfully detecting 93.8% of the cancer cases by manual inspection of LDCT scans using the same radius-based criteria [2].
Despite its high sensitivity, our system is still not robust enough for unsupervised cancer screening. Namely, detecting FP, as showed in Fig. 4C, combined with the simplistic diameter decision which, although diagnostic relevant, is not sufficient to perform cancer prediction because our specificity is only 23.8%. This is in line with the NLST study, where 96.4% of the cancer cases referred for follow-up were actually FP detections. Instead, our method is to be used as an independent observer during the screening process since, unlike end-to-end deep learning approaches such as the Kaggle solutions, it has a human understandable reasoning behind the referral decision, i.e., the clinician can verify the structure that the model considered to be of interest for the decision process. This means that our FP predictions can be easily checked by the expert during the screening.
4 Conclusions
We propose and validate a second opinion computer-aided lung cancer screening system that achieves high patient referral sensitivity. Despite its high performance, the decision based on the NLST criteria does not allow the system to be robust in terms of cancer risk prediction. Future research should thus focus on the development of advanced methods capable of characterizing nodules using a single or multiple time-points and predict nodule malignancy accordingly.
Ultimately, the high performance and explainability of our approach makes it an objective second-opinion system for clinicians to use during the screening process and can contribute to further increase the early detection of lung cancer.
References
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2018. CA A Cancer J. Clin. 68(1), 7–30 (2018)
The National Lung Screening Trial Research Team: Reduced lung-cancer mortality with low-dose computed tomographic screening. New England J. Med. 365(5), 395–409 (2011)
Torre, L.A., Siegel, R.L., Ward, E.M., Jemal, A.: Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol. Biomark. Prev. 25(1), 16–27 (2016)
Setio, A., Traverso, A., de Bel, T.: Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med. Image Anal. 42, 1–13 (2017)
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39(6), 1137–1149 (2017)
Messay, T., Hardie, R.C., Tuinstra, T.R.: Segmentation of pulmonary nodules in computed tomography using a regression neural network approach and its application to the lung image database consortium and image database resource initiative dataset. Med. Image Anal. 22(1), 48–62 (2015)
Wang, S., Zhou, M., Liu, Z., et al.: Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med. Image Anal. 40(3), 172–183 (2017)
Armato, S.G., McLennan, G., Bidaut, L.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38(2), 915 (2011)
Redmon, J., Farhadi, A.: YOLO9000: Better, faster, stronger. In: Proceedings of 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, pp. 6517–6525 (2017)
Szegedy, C., Liu, W., Jia, Y., et al.: Going deeper with convolutions. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1–9. IEEE, 7–12 June 2015
Ding, J., Li, A., Hu, Z., Wang, L.: Accurate pulmonary nodule detection in computed tomography images using deep convolutional neural networks, pp. 1–9. arXiv, June 2017
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Acknowledgements
Guilherme Aresta is funded by the FCT grant contract SFRH/BD/120435/2016. Teresa Araújo is funded by the FCT grant contract SFRH/BD/122365/2016. This study is associated with project NLST-375 and LNDetector, which is financed by the ERDF - European Regional Development Fund through the Operational Programme for Competitiveness - COMPETE 2020 Programme and by the National Fundus through the Portuguese funding agency, FCT - Fundação para a Ciência e Tecnologia within project POCI-01-0145-FEDER-016673.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Aresta, G. et al. (2018). Towards an Automatic Lung Cancer Screening System in Low Dose Computed Tomography. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_31
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_31
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)