Abstract
Deep learning methods have been widely and successfully applied to the medical imaging field. Specifically, fully convolutional neural networks have become the state-of-the-art supervised segmentation method in a variety of biomedical segmentation problems. Two fully convolutional networks were proposed to sequentially achieve accurate lobar segmentation. Firstly, a 2D ResNet-101 based network is proposed for lung segmentation and 575 chest CT scans from multicenter clinical trials were used with radiologist approved lung segmentation. Secondly, a 3D DenseNet based network is applied to segment the 5 lobes and a total of 1280 different CT scans were used with radiologist approved lobar segmentation as ground truth. The dataset includes various pathological lung diseases and stratified sampling was used to form training and test sets following a ratio of 4:1 to ensure a balanced number and type of abnormality present. A 3D CNN segmentation model was also built for lung segmentation to investigate the feasibility using current hardware. Using 5-fold cross validation a mean Dice coefficient of 0.988 ± 0.012 and Average Surface Distance of 0.562 ± 0.49 mm was achieved by the proposed 2D CNN on lung segmentation. 3D DenseNet on lobar segmentation achieved Dice score of 0.959 ± 0.087 and Average surface distance of 0.873 ± 0.61 mm.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
1.1 Conventional Lung and Lobar Segmentation
Segmentation of pathologic lungs on CT images has been investigated in a number of studies. Voxel classification utilizing local texture features has been used to segment abnormal lung regions [1]. Applying anatomical model constraint was another strategy to overcome the challenge of pathological lung segmentation [2]. For example, curvature of ribs [3] was used to assist the selection of optimal thresholds to segment the lung. A modified convex hull algorithm [4] was introduced to extract coarse lung regions present with diffuse lung disease followed by morphological analysis as post-processing. Traditional lobar segmentation is usually achieved by fissure detection/segmentation and a variety of methods have been proposed to accomplish this task, such as watershed, level set, and SVM based. Occasionally, segmentation of airway and vessels are used to assist. However, most of the methods were evaluated on a relatively small datasets and their efficiency is not ideal for high throughput processing. More importantly, they still struggle when confronted with pathological lungs which are common in clinical practice.
1.2 Deep Learning in Medical Image Segmentation
Deep learning methods have been successfully applied to various medical image analysis problems [5,6,7]. More specifically, fully convolutional neural networks (FCN) [8] have become the state-of-the-art approach for segmentation on many imaging modalities. U-Net [9] has demonstrated success in many biomedical image segmentation problems, including cell segmentation in microscopic images. SegNet [10], which is a deep Encoder-Decoder neural network, showed promising results regarding lung segmentation in chest radiographs. Volumetric image based methods were also introduced, such as 3D U-Net [11] and V-Net [12], to segment prostate and kidney in MR. As for FCN application on chest CT, Harrison et al. [13] proposed a progressive and multi-path holistically nested 2D network (P-HNN) for lung segmentation. The method was evaluated on a large dataset including many pathological lungs. A V-Net [14] like architecture was proposed by IBM research aimed at volumetrically segmenting the lung with more spatial context. It demonstrated promising results using a 3D CNN. A relatively small input size was used to compromise the memory constraints and this actually caused noticeable under-segmentation near the boundary. George et al. [15] applied the same P-HNN method on lobar segmentation using 2D axial slices, and the crude segmentation from CNN was followed by a 3D random walker to refine it. Presumably, it is difficult to segment different lobes based on 2D slices without spatial context and a 3D volume based method has the potential to overcome this obstacle.
1.3 ResNet and DenseNet
The Residual Network (ResNet [16]) has been the state-of-the-art image recognition architecture and won first place on the ILSVRC-2015 classification task. Due to its much deeper network with residual learning it demonstrated exceptional performance, surpassing previous models such as VGG [17] and GoogleNet [18]. More recently, DenseNet [19] was introduced in which each layer is connected to every other layer in a feed-forward fashion. It is easier to train due to the improved flow of gradients throughout the network and able to achieve better performance using fewer parameters.
In this paper, we will introduce two fully convolutional networks applied sequentially achieve accurate lobar segmentation. In the first stage, a 2D ResNet-101 based model is used for lung segmentation. Thereafter, a 3D DenseNet based network is applied to perform lobar segmentation based on the initial lung segmentation.
2 Materials
The chest CT images used in this study were collected retrospectively from 6 multicenter clinical trials. This provides image acquisition at different sites with variations in slice thickness, reconstruction kernel, scanner, etc. The slice thickness range is [0.625 mm, 3 mm], in-plane (x-y) spacing range is [0.467 mm, 1 mm], tube current range is [80 mA, 644 mA] and reconstruction kernels include a range from smooth to sharp. In total, 575 chest CT scans from different subjects were used, each with radiologist-approved lung segmentation. 143 scans are from subjects enrolled in Chronic Obstructive Pulmonary Disease (COPD) clinical trials and 432 are from interstitial pulmonary fibrosis (IPF) trials. These cases contain common lung parenchymal abnormalities including emphysema, ground glass, fibrosis, nodule, and honeycombing. Sample axial images of these disease patterns are shown in Fig. 1.
The ground truth reference lung segmentations of these scans were derived using an independent semi-automated segmentation. Specifically, the scans were segmented using a threshold-based anatomical model technique [20, 21], followed by manual editing by lab technologists. Finally, radiologists performed review, editing as needed, and approved the final segmentation. Two thoracic radiologists were involved, both with more than 20 years experience.
Balanced sampling, rather than random sampling, was used to form training and test sets that were balanced in terms of the number and type of abnormalities present. The whole image set was sorted based on the mean Hounsfield Unit (HU) within the lung in ascending order. As a consequence, scans with lower and higher mean lung intensity are likely corresponding to cases with emphysema and IPF, respectively. For every 5 scans of the sorted image set, they were split into training, and test following a ratio of 4:1. 5-fold cross validation with balanced distribution was used and each scan was used for testing exactly once. At each fold, the training set consisted of 460 scans, and the test set was composed of 115 scans. Using this approach similar to stratified cross validation [22], the composition distributions of training, and test set are similar to the original dataset. For a specific scan, only slices (axial images) with lung segmentation (i.e., containing lung) were used.
For lobar segmentation, 705 more scans were used to increase the size of data set with a total number of 1280 scans and all of them have corresponding radiologist approved lobar segmentations. The resolution (z spacing) limit is <= 3 mm since our method is 3D volume based. Similar to the previous 575 scans used in lung segmentation, the additional 705 scans also include different types of lung abnormalities. In terms of breath-hold type, 1193 scans were acquired at Total Lung Capacity (TLC) and 87 at Residual Volume (RV). The same data sorting and splitting strategy was used to form training and test set.
3 Methods
3.1 Network Architectures
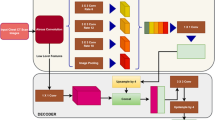
Figure 2 shows the general structure of ResNet-101 based segmentation architecture with input and output size of 256 × 256. The original input image was progressively scaled down 5 times (from 256 × 256 to 8 × 8). Each down-sampling stage includes a residual block, stride of 2 convolution to downsize feature maps. In the de-convolution section, corresponding 5 times up-sampling was used to recover the same resolution as the input image, i.e., from 8 × 8 back to 256 × 256. Each up-sampling stage includes transposed convolution, concatenation and a residual block. Following the same strategy suggested by Szegedy et al. [23], the number of feature maps in the de-convolutional layer was cut by half every time the image size doubles. Also, skip-connections were used in the up-sampling process to incorporate finer details from the lower layers as well as abstract and sematic information from higher layers.
The 3D DenseNet based network is shown in Fig. 3. The down-sampling path includes 4 Transitions Down and 4 Dense Block. Corresponding up-sampling path includes 4 Transitions Up and 4 Dense Block. Transitions Down modules include [Convolution3D, Dropout and Max-pooling]. Transitions Up modules include [Transposed Convolution3D and Dropout]. The Dense Block includes 4 densely connected layers. This architecture was used both in 3D lung segmentation (to compare with 2D CNN) and lobar segmentation. In the case of lung segmentation, the input was a raw image and a sigmoid is used in the final layer. For the lobar segmentation, the input is raw image plus lung mask and softmax is used. To mitigate memory constraints, we decided to perform lobar segmentation on the left and right lung separately. A previous anatomical model based method [21] was used for left and right lung separation.
3.2 Pre-processing, Data Augmentation, and Training
Preprocessing of each CT scan prior to inputting to the CNN involved normalization and rescaling. Image intensity was clipped to range from −1000 HU to +1000 HU and then normalized to [0.0, 1.0]. For 2D model, each slice was resized to 256 × 256.
Similarly, each scan was resized to 128 × 128 × 128 as the input for 3D model. The model was trained from scratch, rather than applying transfer learning using pre-trained weights. Real-time data augmentation was applied using rotation and translation, with the CPU responsible for generating new samples and the GPU for the computation to improve the uniqueness of the augmented data. Dice loss was used as the loss function since it has been shown to be more robust in segmentation problems.
Training of the model was performed using a NVIDIA TITAN X with 12 GB of memory. The Keras deep learning package [24] was used for implementation. Dice Similarity Coefficient and Average Surface Distance were used as evaluation metric.
4 Results
4.1 Lung Segmentation
During testing, the whole scan was fed into the segmentation network to generate a 3D lung segmentation for both the 2D and 3D CNN model. For a single test scan with 200 slices, it takes about 5 s to segment the whole lung.
Table 1 illustrates the segmentation results by the proposed 2D CNN model, 3D CNN model and also previous threshold and anatomical based method [20, 21] developed by our group. Specifically, only 3 out of the total 575 scans from 2D CNN segmentation and 9 scans from 3D CNN segmentation had a Dice coefficient below 0.95. While for our previous method, there were 111 cases with a Dice score below 0.95 from.
Figure 4 shows three example segmentations with different amount of fibrosis present (from mild to severe) by the proposed 2D based CNN, 3D based CNN and previous threshold anatomical model based methods. The second row corresponds to Dice scores of (0.954, 0.931, 0.847) and ASD of (0.575 mm, 1.689 mm, 4.51 mm) by the threshold and anatomical model based method. 2D CNN achieved Dice scores of (0.989, 0.980, 0.976) and ASD of (0.36 mm, 0.505 mm, 1.18 mm). 3D CNN achieved Dice scores of (0.979, 0.969, 0.970) and ASD of (0.484 mm, 0.677 mm, 1.653 mm). The CNN based methods achieved much better segmentation when the attenuation of the lung changed significantly due to disease.
Figure 5 shows example segmentations of emphysema, ground glass and honeycombing by the proposed the CNN methods as well as our previous threshold and anatomical model based method for comparison. In these examples, there are no major attenuation changes in the lung and all three methods were able to achieve good segmentation relative to the ground truth. The Dice scores of these 3 cases (from left to right) are (0.992, 0.973, 0.974) by the threshold and anatomical based method, (0.991, 0.984, 0.976) by 2D CNN, (0.989, 0.981, 0.972) by 3D CNN. Comparable ASD were achieved by the 3 segmentation methods: (0.355 mm, 0.55 mm, 0.503 mm) vs. (0.352 mm, 0.327 mm, 0.479) vs. (0.356 mm, 0.374 mm, 0.488 mm). One minor difference is the successful exclusion of airway by CNN in this honeycombing case.
4.2 Lobar Segmentation
The overall Dice score across 5 lobes is 0.959 ± 0.087 and Average surface distance is 0.873 ± 0.61 mm. More specific performance on each lobe is shown in Table 2.
Figures 6, 7, and 8 show three lobar segmentation examples by sagittal plane: an emphysema case with TLC, a fibrosis case with TLC and a scleroderma case with RV. Detailed Dice score and ASD were shown in their captions respectively.
5 Discussion
The CNN based method achieved highly accurate lung segmentation based on Dice score and Average surface distance. The 2D based CNN model has been successfully applied to segment over 5000 chest CT scans in clinical practice. Our method showed substantial advantages when large amounts of fibrosis are present, especially in peripheral areas close to other soft tissue. Another strength of the CNN based method is its ability to consistently exclude airway trees regardless of the disease patterns and many other methods occasionally require post-processing. In comparison with P-HNN, our work differs in terms of using a radiologist edited and approved reference segmentation for training and testing, as well as the use of a simpler single channel architecture (train from scratch) and no post-processing (such as 3D hole filling).
Using current hardware in clinical practice, the 2D slice based CNN slightly outperformed the 3D volume based CNN. Although 3D based model is able to incorporate more contextual information, this is actually not surprising considering following. Firstly, the variation of slice spacing along the z direction is much larger than in the x-y plane. One strategy would be to resample all the scans to the same resolution but this is computationally expensive. Secondly, the number of training samples is also much smaller when a 3D scan is used instead of individual 2D slices. As such, the 2D model was built with more diverse dataset. Lastly, the image was down-sampled more aggressively in the scenario of 3D CNN (512 to 128 vs. 512 to 256). This would inevitably cause more information loss especially fine details near boundary and this is also observed in the work by IBM research [14].
Using the initial lung segmentation from ResNet-101, the 3D DenseNet model successfully segmented the five lobes correctly on pathological lungs. Conventional fissure detection based methods often fail when fissures are incomplete or impacted by lung abnormalities, such as emphysema, fibrosis. However, our 3D CNN model demonstrated robustness in those challenging cases. One deficiency of our current model is the assumption that five lobes are present which may not be true when a lobe collapses, for example. In the future, we will include these highly abnormal cases in training and also expand our evaluation to include public data sets.
6 Conclusion
We developed two fully convolutional neural network to segment lung and lobes sequentially. The 2D ResNet-101 based segmentation architecture was successfully applied to lung segmentation on chest CT without the need for any additional post-processing. It achieved high accuracy even in the presence of challenging diffuse lung diseases. The 3D DenseNet based network demonstrated competitive lobar segmentation performance on various pathological lungs.
References
Mansoor, A., et al.: A generic approach to pathological lung segmentation. IEEE Trans. Med. Imaging 33(12), 2293–2310 (2014)
Birkbeck, N., et al.: Lung Segmentation from CT with Severe Pathologies Using Anatomical Constraints. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 804–811. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10404-1_100
Prasad, M.N., et al.: Automatic segmentation of lung parenchyma in the presence of diseases based on curvature of ribs. Acad. Radiol. 15(9), 1173–1180 (2008)
Pulagam, A.R., Kande, G.B., Ede, V.K.R., Inampudi, R.B.: Automated lung segmentation from HRCT scans with diffuse parenchymal lung diseases. J. Digit. Imaging 29(4), 507–519 (2016)
Antony, J., McGuinness, K., Connor, N.E.O., Moran, K.: Quantifying radiographic knee osteoarthritis severity using deep convolutional neural networks, arXiv cs.CV, vol. 9, p. 02469 (2016)
Kim, E., Corte-Real, M., Baloch, Z.: A deep semantic mobile application for thyroid cytopathology. In: Proceedings of SPIE, vol. 9789, pp. 97890A–97890A-9 (2016)
Suk, H.-I., Shen, D.: Deep learning-based feature representation for AD/MCI classification. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013. LNCS, vol. 8150, pp. 583–590. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-40763-5_72
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 12 June 2015, vol. 07, pp. 3431–3440 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Kalinovsky, A., Kalinovsky, A., Kovalev, V.: Lung image segmentation using deep learning methods and convolutional neural networks lung image segmentation using deep learning methods and convolutional neural networks, October 2016
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation, arXiv Prepr. arXiv:1606.06650 (2016)
Milletari, F., Navab, N., Ahmadi, S.-A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation, arXiv Prepr. arXiv:1606.04797, pp. 1–11 (2016)
Harrison, A.P., Xu, Z., George, K., Lu, L., Summers, R.M., Mollura, D.J.: Progressive and multi-path holistically nested neural networks for pathological lung segmentation from CT images, CoRR, vol. abs/1706.0 (2017)
Negahdar, M., Beymer, D.: Automated volumetric lung segmentation of thoracic CT images using fully convolutional neural network, February 2018
George, K., Harrison, A.P., Jin, D., Xu, Z., Mollura, D.J.: Pathological pulmonary lobe segmentation from CT images using progressive holistically nested neural networks and random walker, ArXiv e-prints, August 2017
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition, CoRR, vol. abs/1512.0 (2015)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: ImageNet Challenge, pp. 1–10 (2014)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the Inception Architecture for Computer Vision (2015)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings - 30th IEEE Conference Computer Vision Pattern Recognition, CVPR 2017, January, vol. 2017, pp. 2261–2269 (2017)
Brown, M.S., et al.: Method for segmenting chest CT image data using an anatomical model: preliminary results. IEEE Trans. Med. Imaging 16(6), 828–839 (1997)
Brown, M.S., et al.: Reproducibility of lung and lobar volume measurements using computed tomography. Acad. Radiol. 17(3), 316–322 (2010)
Kohavi, R.: A study of cross-validation and bootstrap for accuracy estimation and model selection, pp. 1137–1143 (1995)
Szegedy, C., et al.: Going deeper with convolutions. In: Computer Vision and Pattern Recognition (CVPR) (2015)
Chollet, F.: “Keras.” GitHub (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, X. et al. (2018). High Throughput Lung and Lobar Segmentation by 2D and 3D CNN on Chest CT with Diffuse Lung Disease. In: Stoyanov, D., et al. Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO BIA TIA 2018 2018 2018. Lecture Notes in Computer Science(), vol 11040. Springer, Cham. https://doi.org/10.1007/978-3-030-00946-5_21
Download citation
DOI: https://doi.org/10.1007/978-3-030-00946-5_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00945-8
Online ISBN: 978-3-030-00946-5
eBook Packages: Computer ScienceComputer Science (R0)