Abstract
Skin lesion segmentation (SLS) in dermoscopic images is a crucial task for automated diagnosis of melanoma. In this paper, we present a robust deep learning SLS model represented as an encoder-decoder network. The encoder network is constructed by dilated residual layers, in turn, a pyramid pooling network followed by three convolution layers is used for the decoder. Unlike the traditional methods employing a cross-entropy loss, we formulated a new loss function by combining both Negative Log Likelihood (NLL) and End Point Error (EPE) to accurately segment the boundaries of melanoma regions. The robustness of the proposed model was evaluated on two public databases: ISBI 2016 and 2017 for skin lesion analysis towards melanoma detection challenge. The proposed model outperforms the state-of-the-art methods in terms of the segmentation accuracy. Moreover, it is capable of segmenting about 100 images of a \(384\times 384\) size per second on a recent GPU.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
According to the Skin Cancer Foundation statistics, the percentage of both melanoma and non-melanoma skin cancers is rapidly being increased over the last few years [18]. Dermoscopy, non-invasive dermatology imaging methods, can help the dermatologists to inspect the pigmented skin lesions and diagnose malignant melanoma at an initial-stage [11]. Even the professional dermatologists can not properly classify the melanoma only by relying on their perception and vision. Sometimes human tiredness and other distractions during visual diagnosis can also yield a high number of false positives. Therefore, a Computer-Aided Diagnosis (CAD) system is needed to assist the dermatologists to properly analyze the dermoscopic images and accurately segment the melanomas. Many melanoma segmentation approaches have been proposed in the literature. An overview on numerous melanoma segmentation techniques is presented in [25]. However, this task is still a challenge, since the dermoscopic images have various complexities including different sizes and shapes, fuzzy boundaries, different colors and the presence of hair [7].
In last few decades, many approaches have been proposed to cope with the aforementioned challenges. Most of them are based on thresholding, edge-based, region-based active contour models, clustering and supervised learning [4]. However, these methods are unreliable when dermoscopic images are inhomogeneous or lesions have fuzzy or blurred boundaries [4]. Furthermore, their performance relies on efficient pre-processing algorithms, such as illumination correction and hair removal, which badly affect the generalizability of these models.
Recently, deep learning methods applied to image analysis, specially Convolutional Neural Networks (CNNs) have been used to solve the image segmentation problem [14]. These CNN-based methods can automatically learn features from raw pixels to distinguish between background and foreground objects to attain the final segmentation. Most of these approaches generally are based on encoder-decoder networks [14]. The encoder networks are used for extracting the features from the input images, in turn the decoder ones used to construct the segmented image. The U-net network proposed in [17] has been particularly designed for biomedical image segmentation based on the concept of Fully Convolutional Networks (FCN) [14]. The U-net model reuses the feature maps of the encoder layers to the corresponding decoders and concatenates them to upsampled decoder feature maps, which are also called “skip-connections”. The U-Net model for SLS outperformed many classical clustering techniques [13].
In addition, the deep residual network (ResNet) model [23] is a 50-layers network designed for segmentation tasks. ResNet blocks are used to boost the overall depth of the networks and allow more accurate segmentation depending on more significant image features. Moreover, Dilated Residual Networks (DRNs) proposed in [22] increase the resolution of the ResNet blocks’s output by replacing a subset of interior subsampling layers by dilation [21]. DRNs outperform the normal ResNet without adding algorithmic complexity to the model. DRNs are able to represent both tiny and large image features. Furthermore, a Pyramid Pooling Network (PPN) that is able to extract additional contextual information based on a multi-scale scheme is proposed for image segmentation [26].
Inspired by the success of the aforementioned deep models for semantic segmentation, we propose a model combining skip-connections, dilated residual and pyramid pooling networks for SLS with different improvements. In our model, the encoder network depends on DRNs layers, in turn the decoder depends on a PPN layer along with their corresponding connecting layers. More features can be extracted from the input dermoscopic images by combining DRNs with PPN, in turn it also enhances the performance of the final network. Finally, our SLS segmentation model uses a new loss function, which combines Negative Log Likelihood (NLL) and End Point Error (EPE) [1]. Mainly, cross-entropy is used for multi-class segmentation models, however it is not as useful as NLL in binary class segmentation. Thus, in such melanoma segmentation, we propose to use NLL as a loss function. In addition, for preserving the melanoma boundaries, EPE is used as a content loss function. Consequently, this paper aims at developing an automated deep SLS model with two main contributions:
-
An encoder-decoder network for efficient SLS without any pre- and post-processing algorithms based on dilated residual and pyramid pooling networks to enclose coarse-to-fine features of dermoscopic images.
-
A new loss function that is a combination of Negative Log Likelihood and End Point Error for properly detecting the melanoma with weak edges.
2 Proposed Model
2.1 Network Architecture
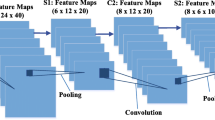
Figure 1 shows the architecture of the proposed SLSDeep model with DRNs [27] and PPN [9]. The network contains two-fold architecture: encoder and decoder. Regarding the encoder phase, the first layer is a \(3\times 3\) convolutional layer followed by \(3\times 3\) max pooling with stride 2.0 that generates 64 feature maps. This layer uses ReLU as an activation and batch normalization to speed-up the training steps with a random initialization. Following, four pre-trained DRNs blocks are then used to extract 256, 512, 1024 and 2048 feature maps, respectively as shown in Fig. 2. The first, third, and fourth DRNs layers are with stride 1.0, in turn the second one is with stride 2.0. Thus, the size of final output of encoder is 1 / 8 of the input image (e.g. in our model, the input image is in \(384\times 384\) and the output feature maps of the encoder is \(48\times 48\)). For global contextual prior, average pooling is used before feeding to fully connected layers in image classification [20]. However, it is not sufficient to extract necessary information from our skin lesion images. Therefore, we do not use average pooling at the end of the encoder and directly fed the output feature maps to the decoder network.
On the other hand, for the decoder network, we use the concept of PPN for producing multi-scale (coarse-to-fine) feature maps and then all scales are concatenated together to get more robust feature maps. PPN use a hierarchical global prior of variant size feature maps in multi-scales with different spatial filters as shown in Fig. 2. In this paper, the used PPN layer extracts feature maps using four pyramid scales with rescaling sizes of \( 1 \times 1, 2 \times 2, 3 \times 3 \) and \(6 \times 6\). A convolutional layer with a \(1 \times 1\) kernel in every pyramid level is used for generating 1024 feature maps. The low-dimension feature maps are then upsampled based on bilinear interpolation to get the same size of the input feature maps. The input and four feature maps are finally concatenated to produce 6144 feature maps (i.e., 4 \(\times \) 1024 feature maps concatenated with the input 2048 feature maps). Sequentially, two \(3\times 3\) convolutional layers are followed by two upsampling layers. Finally, a softmax function (i.e. normalized exponential function) is utilized as logistic function for producing the final segmentation map. A ReLU activation with batch normalization is used in the two convolutional layers [10]. Moreover, in order to avoid the overfitting problem, the dropout function with a ratio of 0.5 [19] is used before the second upsampling layer.
The skip connections between all layers of the encoder and decoder were tested during the experiments. However, the best results were provided when only one connection was skipped between the last layer of the encoder and the output of PPN layer of the decoder. The details of the encoder and decoder architectures are given in the supplementary materials.
2.2 Loss Function
Most of the traditional deep learning methods commonly employ cross-entropy as a loss function for segmentation [17]. Since the melanoma is mostly a small part of a dermoscopic image, the minimization of cross-entropy tends to be biased towards the background. To cope with this challenge, we propose a new loss function by combining objective and content losses: NLL and EPE, respectively. In order to fit a log linear probability model to a set of binary labeled classes, the NLL that is our objective loss function is minimized.
Let \( v \in \{0,1\}\) be a true label for binary classification and \( p = {Pr}(v = 1)\) a probability estimate, the NLL of the binary classifier can be defined as:
In order to maximize Peak Signal-to-Noise Ratio, a content loss function based on an end-point error proposed in [1] is used for preserving the melanoma boundaries. In EPE, We compared the magnitude and orientation of the edges of the predicted mask with the correct one. Let M a generated mask and G the corresponding ground-truth, then the EPE can be defined as:
where (\(M_x\), \(M_y\)) and (\(G_x\), \(G_y\)) are the first derivatives of M and G, respectively in x and y directions.
Thus, our final loss function combining the NLL and EPE can be defined as:
where \(\alpha < 1\) is a weighted coefficient. In this work, we use \(\alpha =0.5\).
3 Experimental Setup and Evaluation
Database: To test the robustness of the proposed model, it was evaluated on two public benchmark datasets of dermoscopy images for skin lesion analysis: ISBI 2016 [6] and ISBI 2017 [8]. The datasets images are captured by different devices at various top clinical centers around the world. In ISBI 2016 dataset, training and testing part contain 900 and 379 annotated images, respectively. The size of the images ranges from \(542\times 718\) to \(2848\times 4288\) pixels. In turn, ISBI 2017 dataset is divided into training, validation and testing parts with 2000, 150 and 600 images, respectively.
Evaluation Metrics: We used the evaluation metrics of ISBI 2016 and 2017 challenges for evaluating the segmentation performances including Specificity(SPE), Sensitivity(SEN), Jaccard index(JAC), Dice coefficient(DIC) and Accuracy(ACC) detailed in [6, 8].
Implementation: The proposed model is implemented on an open source deep learning library named PyTorch [15]. For optimization algorithm, we used Adam [12] for adjusting learning rate, which depends on first and second order moments of the gradient. We used a “poly” learning rate policy [5] and selected a base learning rate of 0.001 and 0.01 for encoder and decoder, respectively with a power of 0.9. For data augmentation, we selected random scale between 0.5 and 1.5, random rotation between −10 and 10 degrees. The“batchsize” is set to 16 for training and the epochs to 100. All the experiments are executed on NVIDIA TITAN X with 12GB memory taking around 20 hours to train the network.
Evaluation and Results: Since the size of the given images is very large, we resized the input images to \(384\times 384\) pixels for training our model. In this work, we tested different sizes and the \(384\times 384\) size yields the best results. In order to separately assess the different contributions of this model, the resulting segmentation for the proposed model with different variations have been computed: (a) The SLSDeep model without the content loss EPE (SLSDeep-EPE), (b) the proposed method with skip connections of all encoder and decoder layers (SLSDeep+ASC) and (c) the final proposed model (SLSDeep) with NLL and EPE loss functions and only one skip connection between the last layer of the encoder and the PPN layer. Quantitative results on ISBI’2016 and ISBI’2017 datasets are shown in Table 1. Regarding ISBI’2016, we compared the SLSDeep and its variations to the four top methods: ExB, [16, 23, 24] providing the best results according to [8]. The segmentation results of our model SLSDeep with its variations (SLSDeep-EPE and SLSDeep+ASC) provided better results than the other four evaluated methods on the ISBI’2016 in terms of the five aforementioned evaluation metrics. SLSDeep yields the best results among the three variations. In addition, for the DIC score, our model, SLSDeep, improved the results with around \(3.5\%\), while the JAC score was significantly improved with \(8\%\). The SLSDeep yielded results with an overall accuracy of more than \(98\%\). Furthermore, SLSDeep on the ISBI’2017 provided segmentation results with improvements of \(3\%\) and \(2\%\) in terms of DIC and JAC scores, respectively. Again SLSDeep outperformed the three top methods of the ISBI’2017 benchmark, [2, 3, 24], in terms of ACC, DIC and JAC scores. However, [24] yielded the best SEN score with just a \(0.9\%\) improvement than our model. The SLSDeep-EPE and SLSDeep+ASC provided reasonable results, however their results were worse than the other tested methods in terms of ACC, DIC, JAC and SEN. However, SLSDeep-EPE yields the highest SPE with a \(0.1\%\) and \(0.3\%\) more than MResNet-Seg [3] and SLSDeep, respectively. Using the EPE function with the final SLSDeep model significantly improved the DIC and JAC scores of \(3\%\) and \(5\%\), respectively, on ISBI’2016 and of \(5\%\) and \(8\%\), respectively, with ISBI’2017. In addition, SLSDeep with only one skip connections yields better results than SLSDeep+ASC on both ISBI datasets.
Qualitative results of four examples from the ISBI’2017 dataset are shown in Fig. 3. For the first and second examples (on the top- and down-left side), the lesions were properly detected, although the color of the lesion area is very similar to the rest of the skin. In addition, the lesion area was accurately segmented regardless the unclear melanoma edges. Regarding the third example (on the top-right side), SLSDeep properly segmented the lesion area; however a small false region having similar melanoma features was also detected. The last example is very tricky, since the lesion shown is very small. However, the SLSDeep model was able to detect it, but with a large false negative region.
Segmentation results: (a) input image, (b) ground truth and (c) correct segmentation by our model, (c’) incorrect segmentation by our model, (d) segmentation by Yuan et al. [24] model.
4 Conclusions
This paper proposed a novel deep learning skin lesion segmentation model based on training an encoder-decoder network. The encoder network used the dilated ResNet layers with downsampling to extract the features of the input image, in turn convolutional layers with pyramid pooling and upsampling are used to reconstruct the segmented image. This approach outperforms, in terms of skin lesion segmentation, the literature evaluated on two ISBI’2016 and ISBI’2017 datasets. The quantitative results show that SLSDeep is a robust segmentation technique based on different evaluation metrics: accuracy, Dice coefficient, Jaccard index and specificity. In addition, qualitative results show promising skin lesion segmentation. Future work aims at applying the proposed model to various medical applications to prove its versatility.
References
Baker, S., Scharstein, D., Lewis, J., Roth, S., Black, M.J., Szeliski, R.: A database and evaluation methodology for optical flow. IJCV 92(1), 1–31 (2011)
Berseth, M.: ISIC 2017-skin lesion analysis towards melanoma detection. arXiv preprint arXiv:1703.00523 (2017)
Bi, L., Kim, J., Ahn, E., Feng, D.: Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. preprint arXiv:1703.04197 (2017)
Celebi, M.E., Wen, Q., Iyatomi, H., Shimizu, K., Zhou, H., Schaefer, G.: A state-of-the-art survey on lesion border detection in dermoscopy images. In: Dermoscopy Image Analysis, pp. 97–129 (2015)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. arXiv preprint arXiv:1606.00915 (2016)
Codella, N.C., et al.: Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). arXiv preprint arXiv:1710.05006 (2017)
Day, G.R., Barbour, R.H.: Automated melanoma diagnosis: where are we at? Skin Res. Technol. 6(1), 1–5 (2000)
Gutman, D., et al.: Skin lesion analysis toward melanoma detection: a challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv preprint arXiv:1605.01397 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. PAMI 37(9), 1904–1916 (2015)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: ICML, pp. 448–456 (2015)
Kardynal, A., Olszewska, M.: Modern non-invasive diagnostic techniques in the detection of early cutaneous melanoma. J. Dermatol. Case Rep. 8(1), 1 (2014)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Lin, B.S., Michael, K., Kalra, S., Tizhoosh, H.: Skin lesion segmentation: U-nets versus clustering. arXiv preprint arXiv:1710.01248 (2017)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of CVPR, pp. 3431–3440 (2015)
Paszke, A., Gross, S., Chintala, S., Chanan, G.: Pytorch (2017)
Rahman, M., Alpaslan, N., Bhattacharya, P.: Developing a retrieval based diagnostic aid for automated melanoma recognition of dermoscopic images. In: Applied Imagery Pattern Recognition Workshop (AIPR), pp. 1–7. IEEE (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2017. CA Cancer J. Clin. 67(1), 7–30 (2017). https://doi.org/10.3322/caac.21387
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
Szegedy, C., et al.: Going deeper with convolutions. In: CVPR (2015)
Yu, F., Koltun, V.: Multi-scale context aggregation by dilated convolutions. arXiv preprint arXiv:1511.07122 (2015)
Yu, F., Koltun, V., Funkhouser, T.: Dilated residual networks. In: Computer Vision and Pattern Recognition, vol. 1 (2017)
Yu, L., Chen, H., Dou, Q., Qin, J., Heng, P.A.: Automated melanoma recognition in dermoscopy images via very deep residual networks. TMI 36(4), 994–1004 (2017)
Yuan, Y., Chao, M., Lo, Y.C.: Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv preprint arXiv:1703.05165 (2017)
Zhang, X.: Melanoma segmentation based on deep learning. Comput. Assist. Surg. 22(sup1), 267–277 (2017)
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: IEEE Conference on CVPR, pp. 2881–2890 (2017)
Zhou, B., Zhao, H., Puig, X., Fidler, S., Barriuso, A., Torralba, A.: Scene parsing through ADE20K dataset. In: Proceedings of the IEEE Conference CVPR (2017)
Acknowledgement
This research is funded by the program Marti Franques under the agreement between Universitat Rovira i Virgili and Fundació Catalunya La Pedrera.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Sarker, M.M.K. et al. (2018). SLSDeep: Skin Lesion Segmentation Based on Dilated Residual and Pyramid Pooling Networks. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham. https://doi.org/10.1007/978-3-030-00934-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-00934-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00933-5
Online ISBN: 978-3-030-00934-2
eBook Packages: Computer ScienceComputer Science (R0)