Abstract
Confocal laser endomicroscopy (CLE) is a novel imaging modality that provides in vivo histological cross-sections of examined tissue. Recently, attempts have been made to develop miniaturized in vivo imaging devices, specifically confocal laser microscopes, for both clinical and research applications. However, current implementations of miniature CLE components such as confocal lenses compromise image resolution, signal-to-noise ratio, or both, which negatively impacts the utility of in vivo imaging. In this work, we demonstrate that software-based techniques can be used to recover lost information due to endomicroscopy hardware miniaturization and reconstruct images of higher resolution. Particularly, a densely connected convolutional neural network is used to reconstruct a high-resolution CLE image, given a low-resolution input. In the proposed network, each layer is directly connected to all subsequent layers, which results in an effective combination of low-level and high-level features and efficient information flow throughout the network. To train and evaluate our network, we use a dataset of 181 high-resolution CLE images. Both quantitative and qualitative results indicate superiority of the proposed network compared to traditional interpolation techniques and competing learning-based methods. This work demonstrates that software-based super-resolution is a viable approach to compensate for loss of resolution due to endoscopic hardware miniaturization.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
Last year, colorectal cancer caused an estimated 50,260 deaths in the United States alone and another 140,030 people are expected to be diagnosed with this disease during 2018 [12, 13]. Accordingly, it is the third most commonly diagnosed cancer among both men and women [13]. Early diagnosis and treatment of colorectal cancer is crucial for reducing the mortality rate. Gastroenterologists screen and monitor the status of their patients’ digestive systems through specialized endoscopy procedures such as colonoscopy and sigmoidoscopy. During colonoscopy, a flexible video endoscope is guided through the large intestine, capturing images used to differentiate between neoplastic (intraepithelial neoplasia, cancer) and non-neoplastic (e.g., hyperplastic polyps) tissues.
Since the introduction of endoscopy to gastroenterology, many significant advances have been made toward improving the diagnostic and therapeutic yield of endoscopy. Confocal laser endomicroscopy (CLE), first introduced to the endoscopy field in 2004 [5], is an emerging imaging modality that allows histological analysis at cellular and subcellular resolutions during ongoing endoscopy. An endomicroscope is integrated into the distal tip of a conventional video colonoscope, providing an in vivo microscopic visualization of tissue architecture and cellular morphology in real-time. Endomicroscopes offer a magnification and resolution comparable to that obtained from ex vivo histology imaging techniques, without the need for biopsy (i.e., tissue removal, sectioning and staining).
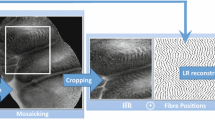
Despite the promise of confocal laser endomicroscopy, both clinicians and researchers prefer compact instruments with relatively large penetration depth to recognize tissue structures such as the mucosa, the submucosa, and the muscular layers. Compact instruments can also directly benefit the patients, as smaller devices improve early diagnostic procedures by offering greater flexibility during hand-held use, for a quicker and less invasive endoscopy [3]. In this regard, further attempts have been made to design miniaturized confocal scanning lasers capable of capturing images from the tissue subsurface with micron resolution in vivo, once installed on top of a flexible fiber bundle. However, miniaturization implies using smaller optical elements, which introduces pixelation artifacts in images. Therefore, there exists a trade-off between miniaturizing the CLE components and the resultant image resolution.
Image super-resolution, transforms an image from low-resolution (LR) to high-resolution (HR) by recovering the high-frequency cues and reconstructing textural information. In the past decade, various learning-based approaches have been proposed to learn the desired LR-to-HR mapping, including dictionary learning [18, 19], linear regression [14, 17], and random decision forests [10].
In recent years, deep learning models have been applied to various image interpretation tasks. Among such efforts, convolutional neural networks (CNN) have been utilized to resolve the ill-posed inverse problem of super-resolution. Dong et al. [1] demonstrated that a fully convolutional network trained end-to-end can be used to perform the LR-to-HR nonlinear mapping. The same authors extended their previous work by introducing deconvolutional layers at the end of the architecture, such that the mapping between LR and HR images is learned directly without image interpolation [2]. They also slightly increased the depth of the network and adopted smaller kernels for better performance. Instead of HR images, Kim et al. [6] suggested to train deeper neural networks through predicting the residual images, which when summed with an interpolated image gives the desired output. Increasing the network depth by adding weighted layers introduces more parameters, which can lead to overfitting. Kim et al. [7] tackled overfitting by using a deeply-recursive convolutional network. In their work, the same convolutional layers are used recursively without the need for extra parameters. To simplify the training of the network, they suggested recursive supervision and skip connections to avoid the notorious vanishing/exploding gradients.
Given the constraints imposed by CLE hardware miniaturization, we propose to leverage state-of-the-art deep learning super-resolution methods to mitigate the unwanted trade-off between miniaturization and image resolution. In other words, we show that the pixelation artifact, which is a consequence of hardware miniaturization, can be significantly remedied through an efficient and practical use of software-based techniques, particularly machine learning methods. To this end, we employ a densely connected CNN in which extensive usage of skip connections is exploited [15]. Dense connections help information flow in backpropagation algorithms and alleviate the vanishing gradient problem. Furthermore, the low-level features from early layers are efficiently combined with those of later layers. In addition, we use sub-pixel convolutional layers [11] to render the upsampling operation learnable and expedite the reconstruction process.
2 Method
Our main goal in this work is to super-resolve an LR image by passing it through a set of nonlinear transformations to recover high-frequency details and reconstruct the HR image, effectively increasing the number of pixels from \(N_{LR}\times N_{LR} \text { to } N_{HR}\times N_{HR}\), where \(\frac{N_{HR}}{N_{LR}}\) is the scale factor. The proposed architecture consists of dense blocks and upsampling layers which are efficiently designed to combine the features from earlier layers with those of later layers and improve information flow throughout the model. Figure 1 depicts the architecture of the employed model.
Low-level Features. A series of low-level features are extracted from small regions of the LR input image using two successive convolutional layers with kernel size \(3\times 3\) and ReLU non-linearity. The number of feature channels for the first and second layer is 64 and 128, respectively. The learned low-level features are used to efficiently represent the intrinsic textural differences between LR and HR images.
High-level Features. The resultant low-level feature maps are used as the input to a fully convolutional DenseNet architecture to provide high-level features. DenseNet, which was first introduced by Huang et al. [4], consists of a set of dense blocks in which any layer is connected to every other layer in a feed-forward fashion. Alternatively stated, the \(i^{th}\) layer in a dense block receives the concatenation of outputs by all preceding layers as the input:

where \(\psi _{\theta ^i}\) denotes the transformation of the \(i^{th}\) layer parameterized by \(\theta ^i\) and  denotes the concatenation operation. Dense skip connections help alleviate the vanishing-gradient problem and improve information flow throughout the network. Counter-intuitively, the number of parameters is also reduced since the previously-generated feature maps are re-used in the subsequent layers, thus minimizing the need for learning redundant features. As depicted in Fig. 1, a single dense block consists of m convolutional layers, each producing k feature maps, referred to as the growth rate. Accordingly, the final output of each dense block has \(m \times k\) features maps. The growth rate regulates how much new information each layer contributes to achieving the final performance. In this study, we set m and k to be 8 and 16, respectively. Thus, each dense block receives and produces 128 feature maps as input and output. We stack 12 dense blocks in a feed-forward fashion to construct the DenseNet part of our proposed architecture.
denotes the concatenation operation. Dense skip connections help alleviate the vanishing-gradient problem and improve information flow throughout the network. Counter-intuitively, the number of parameters is also reduced since the previously-generated feature maps are re-used in the subsequent layers, thus minimizing the need for learning redundant features. As depicted in Fig. 1, a single dense block consists of m convolutional layers, each producing k feature maps, referred to as the growth rate. Accordingly, the final output of each dense block has \(m \times k\) features maps. The growth rate regulates how much new information each layer contributes to achieving the final performance. In this study, we set m and k to be 8 and 16, respectively. Thus, each dense block receives and produces 128 feature maps as input and output. We stack 12 dense blocks in a feed-forward fashion to construct the DenseNet part of our proposed architecture.
Upsampling Layers. In some SR methods [1, 6, 16], the LR image is first resized to match the HR spatial dimensions using bicubic interpolation. Thereafter, several convolution layers are employed to enhance the interpolated input in the HR space. In addition to having a considerable increase in memory usage and computational complexity, these interpolation methods are categorized as non-learnable upsampling techniques, which do not leverage data statistics to bring new information for more accurate reconstruction. As an alternative, deconvolutional layers, which are learnable operations, are utilized to enlarge the spatial dimensions of the LR image. However, the most prominent problem associated with deconvolutional layers is the presence of checkerboard artifacts in the output image. To overcome this, extra post-processing steps or smoothness constraints are required. In this work, we use sub-pixel convolutional layers [11], to upsample the spatial size of the feature maps within the network. Suppose that we desire to spatially upsample c feature maps of size \(h \times w \times c\) to size \(H \times W \times c\), by a scale factor \(r=H/h=W/w\). The LR feature maps would be fed into a convolution layer that increases the number of channels by a factor of \(r^2\), resulting in a volume of size \(h \times w \times (c \times r^2)\). Next, the resultant volume is simply re-arranged to be of shape \((h\times r) \times (w \times r) \times c\), which is equal to \(H \times W \times c\). Here, we use successive \(\times 2\) upsampling layers to gradually increase the spatial dimensionality. Each upsampling block contains a single convolutional layer with \(3 \times 3\) kernel size and ReLU non-linearity.
Integration Layer. Once the features maps match the spatial dimension in the HR space, an integration layer is used to consolidate the features across the channels into a single channel. The integration layer is a convolutional layer with \(3 \times 3\) kernel size and a single output channel. Finally, a sigmoid activation function is employed to produce the super-resolved image.
3 Experiments
Data. We evaluate our study on the dataset provided by Leong et al. [9]. The dataset contains 181 gray scale confocal images of size \(1024 \times 1024\) from 31 patients and 50 different anatomical sites. Each patient has undergone a confocal gastroscopy (Pentax EC-3870FK, Pentax, Tokyo, Japan) under conscious sedation. CLE images and forceps biopsies of the same sites were taken sequentially at standardized locations (i.e., sites of the small intestine). Each forceps biopsy was then assessed by 2 experienced blinded histopathologists. Despite our application of interest being colorectal cancer, we used the publicly available CLE celiac dataset as a proof-of-concept. Colorectal cancer images are assessed primarily in the large intestine as opposed to the small intestine used in celiac assessment, however the imaging procedure (CLE) remains the same. This dataset was made publicly available as part of an International Symposium on Biomedical Imaging (ISBI) challenge and we used the provided training and test sets, consisting of 108 and 73 images, respectively.
Implementation Details. We partition the HR images into \(64\times 64\) non-overlapping patches. Then, the HR patches are downsampled by bicubic interpolation to construct <LR, HR> pairs for training the model. The network is optimized with Adam [8] optimizer with default parameters, i.e. \(\beta _1=0.9\), \(\beta _2=0.999\) and \(\epsilon =10^{-4}\). We set the mini-batch size to 128. The learning rate is first initialized with 0.001 and is multiplied by \(\gamma =10\) at epochs 50 and 200. The network is trained for 300 epochs using L1 loss. For data augmentation, we use random horizontal and vertical flips. The proposed method is implemented in PyTorch and is trained using two Nvidia Titan X (Pascal) GPUs. It takes 2 days to train the networks for each upsampling factor. All hyper-parameters (optimizer, learning rate, batch size, and distance metric) are found via grid search on 20 images from the training set.
Qualitative Results. In Fig. 2, we visually compare our proposed super-resolution method to three traditional interpolation techniques and two learning-based approaches with scale factors of \(\times 2\), \(\times 4\) and \(\times 8\). Evidently, DenseNet produces output images of higher quality by reconstructing high-frequency cues and removing visual artifacts, e.g. over-smoothness and pixelation. Specifically for a \(\times 8\) scale factor, the densely connected network can accurately recover high-level textural patterns such as grids and granular patterns. Moreover, a more rigorous examination of smaller regions for \(\times 4\) scale factor clearly reveals the superiority of DenseNet model in producing sharper edges and improved contrast for lines and shapes.
From a clinician’s point of view, the reconstruction power of the method offers a clear advantage over others. In Fig. 3 we illustrate the trade-off between the amount of lost information after downsampling and the quality of the reconstructed image. As can be seen, a large portion of pixels is discarded in downsampling, restricting the networks to a small fraction of the original image pixels for reconstruction. However, deep learning approaches are clearly capable of generating a sharp image from only 1.6% of pixels (for a scale factor of \(\times 8\)) with very small L1 distance values which indicates a minimal loss of information.
Quantitative Results. Table 1 compares our proposed method with three interpolation methods and two learning-based techniques in terms of PSNR (Peak Signal to Noise Ratio) and SSIM (Structural Similarity). PSNR is a well-known metric for image quality assessment which is inversely proportional to Mean Square Error. SSIM also measures the the similarity between two images and is correlated with quality perception in human visual system. In terms of PSNR, DenseNet yields 2.08, 1.93 and 1.14 average improvements over Nearest, Bilinear and Bicubic interpolation methods across all scale factors, respectively. For learning-based approaches, DenseNet outperforms A+ [14] and SRCNN [1] in terms of average SSIM by 0.020 and 0.019 over all scale factors, respectively.
4 Conclusion
Developing smaller hardware for medical imaging devices has several advantages such as increased portability and reduced patient discomfort. However, hardware miniaturization comes at the expense of reduced image quality. In this preliminary study, we obtained encouraging results to support that software-based methods can be used to counteract the loss of image quality due to miniaturized device components. Compared to common interpolation methods, our qualitative and quantitative results indicate that a densely connected convolutional neural network can significantly yield higher PSNR and SSIM scores, resulting in super-resolved images of higher quality. In future work, we will focus on how super-resolved images, compared to low-resolution images, can be advantageous to clinical and research applications. For example, super-resolution images may be used as input to automated machine-learning based disease classification.
References
Dong, C., et al.: Image super-resolution using deep convolutional networks. IEEE PAMI 38(2), 295–307 (2016)
Dong, C., Loy, C.C., Tang, X.: Accelerating the super-resolution convolutional neural network. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016, Part II. LNCS, vol. 9906, pp. 391–407. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46475-6_25
Helmchen, F.: Miniaturization of fluorescence microscopes using fibre optics. Exper. Physiol. 87(6), 737–745 (2002)
Huang, G., et al.: Densely connected convolutional networks. In: IEEE CVPR, pp. 2261–2269 (2017)
Kiesslich, R., et al.: Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo. Gastroenterology 127(3), 706–713 (2004)
Kim, J., et al.: Accurate image super-resolution using very deep convolutional networks. In: IEEE CVPR, pp. 1646–1654 (2016)
Kim, J., et al.: Deeply-recursive convolutional network for image super-resolution. In: IEEE CVPR, pp. 1637–1645 (2016)
Kingma, D.P., et al.: Adam: A method for stochastic optimization. arXiv:1412.6980 (2014)
Leong, R.W., et al.: In vivo confocal endomicroscopy in the diagnosis and evaluation of celiac disease. Gastroenterology 135(6), 1870–1876 (2008)
Schulter, S., et al.: Fast and accurate image upscaling with super-resolution forests. In: IEEE CVPR, pp. 3791–3799 (2015)
Shi, W., et al.: Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In: IEEE CVPR, pp. 1874–1883 (2016)
Siegel, R.: Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67(3), 177–193 (2017)
Siegel, R.: Cancer statistics, 2018. CA Cancer J. Clin. 68(1), 7–30 (2018)
Timofte, R., De Smet, V., Van Gool, L.: A+: adjusted anchored neighborhood regression for fast super-resolution. In: Cremers, D., Reid, I., Saito, H., Yang, M.-H. (eds.) ACCV 2014, Part IV. LNCS, vol. 9006, pp. 111–126. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-16817-3_8
Tong, T., et al.: Image super-resolution using dense skip connections. In: IEEE ICCV, pp. 4809–4817 (2017)
Wang, Z., et al.: Deep networks for image super-resolution with sparse prior. In: IEEE ICCV, pp. 370–378 (2015)
Yang, C.Y., et al.: Fast direct super-resolution by simple functions. In: IEEE ICCV, pp. 561–568 (2013)
Yang, J., et al.: Image super-resolution via sparse representation. IEEE TIP 19(11), 2861–2873 (2010)
Yang, J., et al.: Coupled dictionary training for image super-resolution. IEEE TIP 21(8), 3467–3478 (2012)
Acknowledgments
Thanks to the NVIDIA Corporation for the donation of Titan X GPUs used in this research and to the Collaborative Health Research Projects (CHRP) for funding.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Izadi, S., Moriarty, K.P., Hamarneh, G. (2018). Can Deep Learning Relax Endomicroscopy Hardware Miniaturization Requirements?. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)