Abstract
Recently, Magnetic Resonance imaging-only (MR-only) radiotherapy treatment planning (RTP) receives growing interests since it is radiation-free and time/cost efficient. A key step in MR-only RTP is the generation of a synthetic CT from MR for dose calculation. Although deep learning approaches have achieved promising results on this topic, they still face two major challenges. First, it is very difficult to get perfectly registered CT-MR pairs to learn the intensity mapping, especially for abdomen and pelvic scans. Slight registration errors may mislead the deep network to converge at a sub-optimal CT-MR intensity matching. Second, training of a standard 3D deep network is very memory-consuming. In practice, one has to either shrink the size of the training network (sacrificing the accuracy) or use a patch-based sliding-window scheme (sacrificing the speed). In this paper, we proposed a novel method to address these two challenges. First, we designed a max-pooled cost function to accommodate imperfect registered CT-MR training pairs. Second, we proposed a network that consists of multiple 2D sub-networks (from different 3D views) followed by a combination sub-network. It reduces the memory consumption without losing the 3D context for high quality CT synthesis. We demonstrated our method can generate high quality synthetic CTs with much higher runtime efficiency compared to the state-of-the-art as well as our own benchmark methods. The proposed solution can potentially enable more effective and efficient MR-only RTPs in clinical settings.
Y. Zhao and S. Hong—This work was mainly accomplished during Yu Zhao, Sungmin Hong’s internship at Siemens Medical Solutions.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Medical imaging plays an important role in radiotherapy treatment planning (RTP) [1] by providing critical information for organ/tumor localization and dose calculation. Currently computed tomography (CT) is the primary modality, which provides electron density information for dose calculation. Since Magnetic Resonance (MR) imaging is more valuable in organ/tumor localization due to its superior soft tissue contrast, it has received more and more interests in RTP. In traditional workflow, MR will be registered to a principal CT dataset [1, 2] so that its superior soft tissue contrast information can be fused with the CT image. However, due to the imperfectness of the current image registration techniques, registration error will bring systematic spatial uncertainty [3], hence, influencing the accuracy of RTP. Recently MR-only RTP receives growing interests since it is radiation-free and time/cost efficient. A key step in MR-only RTP is the generation of a synthetic CT (sCT) from MR for dose calculation.
The major challenge in CT synthesizing is the intensity ambiguity of different tissues, such as bone and air which both appear dark on MR. Traditional approaches for CT synthesis from MR can be divided into two categories: atlas-based [4] and segmentation-based [5]. For the atlas-based approaches, the focus is to register the MR atlas to the patient MR, and then apply the registration transformation on the corresponding CT atlas to generate the synthetic CT [6]. Segmentation-based methods [5] segment different types of tissues from MR. A synthetic CT is then generated by filling a constant CT intensity for each type of tissue. The main obstacles for these approaches are the synthesis speed and registration or segmentation accuracy. Recently, some context-aware deep learning based models are proposed [7,8,9] and they achieved promising results. However, they still face two major challenges. First, standard deep learning requires a set of perfectly registered CT-MR pairs to learn the intensity mapping from MR to CT. However, since MR and CT images are acquired at different time with different patient positionings and table shapes, it is very difficult to perfectly register them, especially for abdomen and pelvic scans [10]. Thus most works [7,8,9] focused on brain regions. Slight registration errors may induce large mis-matching in the intensity space, hence, misleading the deep network to converge at a sub-optimal CT-MR intensity matching. Second, training of a standard 3D deep network is very memory-consuming. In practice, even with a high-end deep learning server, one has to simplify the 3D network structure or using a patch-based sliding-window scheme [7, 9] to accommodate large volumes of training data. The simplified network may not model the MR-CT intensity mapping well and sliding-window scheme may sacrifice the speed significantly.
In this work, we proposed a novel method to tackle the aforementioned challenges. First, we designed a maxpooling loss function allowing the network to search optimal intensity matching not only between the corresponding CT-MR patches but across their neighborhood. This kind of “matching freedom” makes the network robust to imperfect CT-MR registration. Second, we proposed a network consisting of multiple 2D sub-networks (from different 3D views) followed by a 3D combination sub-network. It dramatically reduces the memory consumption without losing the 3D context for high quality CT synthesis. Our method generated high quality sCTs with much higher runtime efficiency compared to the state-of-the-art and our own benchmark method.
2 Materials and Methods
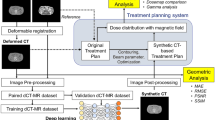
2.1 Overview of Multi-view Multi-channel U-Net Structure
U-Net [11] is a deep network originally proposed for image segmentation. It has a symmetric hierarchical structure that enables precise voxel-wise classification by modeling cross-scale anatomical context. In our study, the U-Net is adapted to a regression network, i.e., the output is an image with synthetic CT values. The original U-Net has a 2D fully convolutional structure, which needs to be extended to handle the 3D nature of MR and CT images. In order to train on 3D volumes without reducing network size and speed, we adopt a 2.5D framework (Fig. 1). Our framework consists of two 2D-centric U-Nets (Fig. 2) corresponding to sagittal and axial views, respectively. The stacked output 3D features from these two sub-nets are further combined by a 3D combination sub-net (Fig. 5). Moreover, to deal with the unpreventable misalignments between MR-CT training pairs for accurate model training, a maxpooling hinge-like Huber function is designed as training loss (Fig. 3). Technical details are explained next.
2.2 Multi-channel MR Inputs for Information Enhancement
The input of our method is the In-Phase and Out-of-phase images generated by MR Dixon method. These images capture complementary fat and water information for tissue differentiation. As shown in Fig. 2, for single view 2D U-Net inputs, instead of stacking these two images at the input layer, we keep two independent channels for each of them. In this way, the network can capture features from different MR sequences independently for information enhancement.
2.3 Maxpooling and Hinge-like Huber Loss Function
In order to learn the intensity transformation from MR to CT, a set of registered MR-CT pairs are needed for training. However, it is very difficult to perfectly register MR and CT due to organ deformations, different table sizes, etc. To address this problem, an effective loss function is proposed for the network in Fig. 2. Instead of calculating the voxel-wise intensity differences directly between the output slice and the ground-truth slice, a maxpooling process (Fig. 3(a)) is applied to accommodate the slight misalignment in a translation invariant fashion [12]. A schematic example is presented in Fig. 4. If I1 and I2 are perfectly registered, it is easy to learn a consistent mapping function F that maps the intensities of pixel a, b and c to aʹ, bʹ and cʹ, i.e., F(I1(a)) = I2 (aʹ), F(I1(b)) = I2 (bʹ), F(I1(c)) = I2 (cʹ); However, if I1 and I2 are not perfectly registered due to deformable or rigid registration errors, it is very difficult to learn a common mapping function that maps the intensities of a, b and c to aʹ, bʹ and cʹ, since the intensity transformation becomes inconsistent. By adding max-pooling in the loss function, we essentially give some spatial freedom to the mapping function, allowing it to map the intensity to its neighborhood, i.e., F(I1(a)) = I2 (aʹ + Δaʹ), F(I1(b)) = I2 (bʹ + Δbʹ), F(I1(c)) = I2 (cʹ + Δcʹ). Thus, a consistent mapping function can be learned. Note that the max-pooling allows different voxels to have different small Δ, which address the non-systematic registration errors. The hinge-like function is also adopted with Huber loss as the final loss function (Fig. 3(b)), also shown in Eq. (1). It accommodates major loss and ignores minor ones.
where a is the 2D image slice difference between output and ground-truth CT images.
A schematic explanation of the impact of mis-registration to intensity transformation. (a) Image 1 (Modality 1), (b) Perfectly registered Image modality 2, (c) Image 2 with rigid mis-alignment, (d) Image 2 with non-rigid mis-alignment. Triangles in (a)–(d) represent the same object. Dashed lines in (c) and (d) denote the locations of the perfectly registered Image 2.
2.4 Multi-view Combination of the 2D U-Net Like Structures
Our network includes two 2D U-Nets followed by a combination network (Fig. 5). This design is important to deal with memory limitations. With an 8 GB GPU memory, we cannot fit a 3D 192 * 224 * 168 volume with a 3D network for training. Therefore, we decompose the 3D volume into 2D axial and sagittal slices, respectively, which can be easily fit into two 2D U-Nets. However, since the 2D U-Net ignores the 3D context across neighboring slides, the output may have stitching blurring effect. (c.f. Fig. 7), To remove the blurring effect, output feature maps of 2D U-Nets are stacked into 3D volumes before feeding into a 3D convolution layer with kernel size \( 1\, \times \,1\, \times \,1 \). This 3D convolution layer effectively removes the 2D stitching blurring effect.
The overall loss function is a (empirically-set) weighted mean of maxpooling hinge-like Huber loss from two views and a Huber loss of the 3D synthetic volume with the ground-truth volume (2).
where \( v \) is the 3D volume difference between the final output synthetic CT of the combination net and the ground-truth CT volumes; 3D \( {\text{L}}\left( v \right)_{sagittal} \) and \( L\left( v \right)_{axial} \) are the hinge-like Huber loss maxpooled from sagittal view and axial view 2D slices respectively; \( Huber\left( v \right) \) is the voxel-wise Huber loss of the 3D volume difference.
2.5 Network Training
Our network training has two stages. First, the two 2D U-Nets of axial and sagittal views are trained independently. Then the feature maps extracted from the second last layers of each 2D U-Net are stacked into 3D volumes and saved as input for further training of the 3D combination network.
3 Results
Due to the lack of perfectly aligned scanned MR and CT pairs, the ground-truth CTs are generated by a multi-atlas-based regression method [13]. The quality of the ground-truth synthetic CT image is confirmed and accepted by experienced oncologists. However, since the multi-atlas-based regression method [13] takes extensive time (i.e., more than 15 min on average) to generate the synthetic CT image, it has limitation in the real world RTP clinical workflow. An Nvidia Quadro M4000 GPU with 8 GB memory was utilized for all the training steps. For the first training stage, training time for each 2D U-Net like structure is dependent on the input size of the images at the corresponding view, 21 h and 95 h for axial view and sagittal view, respectively. For the second training stage, combination net, 7 h was taken. A total time of 123 h was used for the 2-stage training procedure. The testing phase only cost less than 8 s for each subject 3D CT volume synthesis.
3.1 Effectiveness of the Proposed Framework
In the experiment, we have 34 MR-CT pairs, where 27 pairs are used for training and the rest 7 pairs for testing. Our proposed method showed significant improvements at 2D slice level compared to the benchmark U-Net structure Fig. 6. The multi-view combination of the 2D U-Net structures also showed effectiveness on removing the 2D slice stitching blurs across the 3D volume and avoided sacrificing synthetic image quality by shrinking the size of 3D training network (Fig. 7). Comparisons between sCTs generated using our proposed method and ground-truth CTs are discussed in the following sections.
3.2 Synthetic CT Quality Improvement
We can clearly see the small misalignment deficits from the multi-atlases-based sCTs [13] used for our training by comparing input MRs in Fig. 8. However, our proposed method will compensate these slight misalignments by predicting both the bone edge and soft tissue actual locations, which outperformed the state-of-art multi-atlas-based algorithm.
3.3 Synthetic CT Evaluation
To quantitatively measure the reliability and accuracy of the synthetic CT outputted by our framework and the ground-truth CT images, Mean Absolute Error (MAE) (also used in [1, 10]) was utilized:
where \( V_{1} , V_{2} \) represent the synthetic CT and ground-truth volumes, and \( N \) represents total number of the voxels.
As for the 7 pure testing cases, the MAE values are very low (average 16.9 HU) (Table 1). Compared to the state-of-art results (average 58 HU in [10] and around 40 HU in most of works [1]), our method achieves higher accuracy. Compared with 3 benchmark U-Net-based methods, the proposed scheme achieved the best performance (Table 1), demonstrating the effectiveness of our specific design. Besides, the stunning CT synthesis speed (less than 8 s) significantly outperformed the state-of-art multi-atlas-based framework used to generate ground-truth synthetic CTs (more than 10 min), which paves the way for applying the proposed framework to real clinical settings.
4 Discussion
In this work, we explored a deep learning framework for CT synthesis from MR. An average MAE of 19.6 HU and ~10 s synthesis speed outperform state-of-the-art methods. It shows the potency of the proposed deep learning framework in cross modality synthesis. Compared to other methods, the proposed method also showed significant improvement in sCT quality. In order to evaluate if our method is sufficient for MR-only RTP, it is important to evaluate the dose calculated from sCTs, which is part of our future work. This work gives us a new insight into tackling imperfect training pairs and 3D network training memory efficiency problem and the superior results also gives the promise to our framework for other applications.
References
Edmund, J.M., Nyholm, T.: A review of substitute CT generation for MRI-only radiation therapy. Radiat. Oncol. 12, 28 (2017)
Edmund, J.M., et al.: SP-0510: dose planning based on MRI as the sole modality: why, how and when? Radiother. Oncol. 115, S248–S249 (2015)
Paulson, E.S., Erickson, B., Schultz, C., Allen Li, X.: Comprehensive MRI simulation methodology using a dedicated MRI scanner in radiation oncology for external beam radiation treatment planning. Med. Phys. 42, 28–39 (2014)
Sjölund, J., Forsberg, D., Andersson, M., Knutsson, H.: Generating patient specific pseudo-CT of the head from MR using atlas-based regression. Phys. Med. Biol. 60, 825–839 (2015)
Delpon, G., et al.: Comparison of automated atlas-based segmentation software for postoperative prostate cancer radiotherapy. Front. Oncol. 6, 178 (2016)
Dowling, J.A., et al.: An Atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int. J. Radiat. Oncol. 83, e5–e11 (2012)
Nie, D., et al.: Medical image synthesis with context-aware generative adversarial networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017, Part III. LNCS, vol. 10435, pp. 417–425. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_48
Wolterink, J.M., Dinkla, A.M., Savenije, M.H.F., Seevinck, P.R., van den Berg, C.A.T., Išgum, I.: Deep MR to CT synthesis using unpaired data. In: Tsaftaris, S.A., Gooya, A., Frangi, A.F., Prince, J.L. (eds.) SASHIMI 2017. LNCS, vol. 10557, pp. 14–23. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-68127-6_2
Nie, D., Cao, X., Gao, Y., Wang, L., Shen, D.: Estimating CT image from MRI data using 3D fully convolutional networks, 1 January 2016
Andreasen, D., et al.: Computed tomography synthesis from magnetic resonance images in the pelvis using multiple random forests and auto-context features. In: Styner, M.A., Angelini, E.D. (eds.) SPIE Medical Imaging, p. 978417. International Society for Optics and Photonics (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015, Part III. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Scherer, D., Müller, A., Behnke, S.: Evaluation of pooling operations in convolutional architectures for object recognition. In: Diamantaras, K., Duch, W., Iliadis, L.S. (eds.) ICANN 2010, Part III. LNCS, vol. 6354, pp. 92–101. Springer, Heidelberg (2010). https://doi.org/10.1007/978-3-642-15825-4_10
Liao, S., et al.: Automatic lumbar spondylolisthesis measurement in CT images. IEEE Trans. Med. Imaging 35, 1658–1669 (2016)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhao, Y. et al. (2018). Towards MR-Only Radiotherapy Treatment Planning: Synthetic CT Generation Using Multi-view Deep Convolutional Neural Networks. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_33
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_33
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)