Abstract
Due to inevitable differences between the data used for training modern CAD systems and the data encountered when they are deployed in clinical scenarios, the ability to automatically assess the quality of predictions when no expert annotation is available can be critical. In this paper, we propose a new method for quality assessment of retinal vessel tree segmentations in the absence of a reference ground-truth. For this, we artificially degrade expert-annotated vessel map segmentations and then train a CNN to predict the similarity between the degraded images and their corresponding ground-truths. This similarity can be interpreted as a proxy to the quality of a segmentation. The proposed model can produce a visually meaningful quality score, effectively predicting the quality of a vessel tree segmentation in the absence of a manually segmented reference. We further demonstrate the usefulness of our approach by applying it to automatically find a threshold for soft probabilistic segmentations on a per-image basis. For an independent state-of-the-art unsupervised vessel segmentation technique, the thresholds selected by our approach lead to statistically significant improvements in F1-score \((+2.67\%)\) and Matthews Correlation Coefficient (+\(3.11\%\)) over the thresholds derived from ROC analysis on the training set. The score is also shown to correlate strongly with F1 and MCC when a reference is available.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
The ability to automatically assess the quality of the outcomes produced by CAD systems when they are meant to work in real clinical scenarios is critical. Unfortunately, internal validation data can be contaminated when used for incremental method development, leading to over-optimistic performance expectations. In addition, differences between the data used for training a model and the data that such model encounters in practice may lead to relevant failures. On the other hand, the availability of automatic quality control tools is key for the effective deployment and monitoring of computational tools on large-scale medical image analysis studies or clinical routines. Unfortunately, this aspect of the CAD system design pipeline is seldom addressed in the literature [12].
In the retinal image analysis field, computational models developed for automatic image understanding are ubiquitous. In this context, a task of particular interest is the analysis of the retinal vessel tree. This involves the study of vascular biomarkers that are of great interest as early indicators of potential diseases, like vessel calibers, tortuosity, and fractal dimension. However, in order to reliably extract such biomarkers, the first step is to extract an accurate binary segmentation of the vessels. For this reason, a large body of research has been dedicated to solve this problem [1, 15]. In comparison, few research has been addressed to the related task of determining the quality of the extracted segmentations.
When the task of retinal vessel segmentation is considered as a binary classification problem, standard metrics like accuracy can be applied to measure performance. Nevertheless, due to the sparsity of the vasculature within the retina, vessel segmentation is a highly imbalanced classification problem, and more appropriate performance estimates like sensitivity, specificity, F1 score, or Matthews Correlation Coefficient, are necessary. More advanced techniques have been also proposed, e.g. [3] or [14]. In both cases, the overall strategy is to allow for some error margin, in order to compensate also for inter-observer differences in the ground-truth generation stage. This is achieved by analyzing the degree of overlap between a manual reference and a segmentation in an adaptive manner, and also penalizing dissimilarities and disconnections on the vessel skeletons.
All the above approaches belong to the category of full-reference quality metrics, for which a ground-truth image is required. In this paper, a no-reference quality score for the automatic assessment of retinal vessel segmentations is introduced for the first time. The proposed method operates in the absence of a reference image. This is achieved by designing a CNN that predicts the similarity between a manual segmentation and a corresponding artificially degraded transformation, as summarized in Fig. 1. This similarity can be considered as an estimate of the degraded segmentations’ quality. The provided experimental results demonstrate that, once trained, the proposed model produces a quality score that correlates well with full-reference quality metrics, and is useful to detect deficient segmentations generated by automatic vessel segmentation techniques.
2 Methodology
In this section we provide a detailed step-by-step technical explanation of the approach proposed to build and train a no-reference retinal vessel quality metric.
2.1 Generating Realistic Degraded Vessel Trees
The first step in our approach is to model an incorrectly segmented vessel tree. The most typical artifacts in this case involve under-segmentations, which often lead to vasculature disconnections and thin vessels vanishing. Another common error source in this case is the presence of the optic disk and retinal lesions, which can be confused with vessels by automatic techniques, leading to over-segmentations.
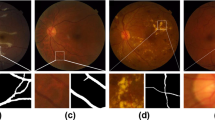
While the latter class of errors is harder to model due to the wide variability of retinal lesions, under-segmentations can be simulated by local morphological erosions, as shown in Fig. 2. We also include morphological dilations, as well as completely white and dark images degraded with impulse noise on the field-of-view, so as to embed in the model information related to over-segmentations.
The examples generated in this stage are produced from a set of manual expert-delineated binary vessel trees, and they are meant to be supplied later to our model in training time. Hence, they need to be generated in an efficient manner. It is also important to train the model with perfect segmentations so that it can correctly attribute a high score in cases where an algorithm successfully separates the vasculature. For this purpose, given a manual vessel tree v, we produce a synthetically degraded version \({\mathbf {deg}}(v)\) of it as follows:
where p is drawn from a uniform probability distribution \(\mathcal {U}(0,1)\), \(\mathcal {N}\) represents impulse noise, and \(\mathcal {M}\) is a stochastic morphological operator that performs a random number of local degradations by first selecting a number n of square image patches \(\bar{v}\) of fixed size, extracted from random location on the original vessel image v. Each of these patches undergoes the following transformations:
where \(\mathcal {E}_s\) and \(\mathcal {D}_s\) are the morphological erosion and dilation operators respectively, specified by a square structuring element s of a size randomly selected to be 3, 5, or 7 at each step. The structuring element is itself randomly built according to a Bernoulli distribution with \(p=0.5\) at each pixel position. Once \(\bar{v}\) has been artificially degraded, it is stored back at its original location in v.
(a) Manual vessel segmentation. (b) Examples of image patches extracted from (a) with corresponding degraded counterparts. (c) Artificially degraded version of (a). The normalized mutual information between (a) and (c) is \(\mathcal {NMI}_{[0,1]}=0.89\). At the patch level, images in the top row of (b) have \(\mathcal {NMI}_{[0,1]}=0.39\), middle row \(\mathcal {NMI}_{[0,1]}=0.23\), and bottom row \(\mathcal {NMI}_{[0,1]}=0.45\).
2.2 Mutual Information as a Similarity Metric Between Binary Vessel Maps
The next step is to establish a similarity measure between a degraded vessel tree and a manually segmented one. Moreover, since the goal is to build an image quality score, this measure should preferably be bounded a-priori in a finite interval. Among the many possibilities, we select the Normalized Mutual Information in order to analyze the amount of shared information in both images.
Given a manually-segmented vessel tree v and its degraded counterpart \({\text {deg}}(v)\), Mutual Information considers both the marginal entropies \(\mathcal {H}(v)\) and \(\mathcal {H}({\text {deg}}(v))\) and the entropy of their joint probability distribution \(\mathcal {H}(v, {\text {deg}}(v))\):
This quantity has been widely used for medical image registration tasks [7]. A derived formulation is the normalized mutual information:
which is more robust to overlaps in both images, and it is bounded as \(1\le \mathcal {NMI} \le 2\), [6]. Finally, in order to obtain a similarity score producing a maximum value of 1 when both images are perfectly aligned, we reparametrize Eq. (4) as follows:
The Normalized Mutual Information formula given by Eq. (5) is suitable for the problem of estimating the similarity of a manual vessel segmentation and a degraded version of it, as shown in Fig. 2.
2.3 A Deep Architecture for Vessel Map Similarity Regression
In order to learn a quality score for binary vessel tree images, we proceed as follows: given a dataset of manually segmented vessel maps, we apply the operator defined in Eq. (1) to build degraded versions, which serve as input for the model. The similarity of the input with its manually-segmented counterpart is computed by means of Eq. (5), which is considered as a proxy to the quality of the degraded vessel tree. For regressing this quantity, we design a CNN as specified below.
The proposed architecture consists of a subsequent application of downsampling and convolution layers, specified in Fig. 3. Downsampling was implemented as a convolution with stride of 2. Non-linear activation functions were applied after each layer, specifically Leaky ReLU units with a slope parameter of \(\alpha =0.02\). A Global Average Pooling (GAP) was applied to obtain a uni-dimensional representation of the input vessel tree of size 512. This representation was supplied to two fully-connected layers, and the output was passed through a sigmoid, resulting in a score in [0, 1]. This score was compared to the previously computed similarity through an \(\mathcal {L}_1\) loss. The weights of the model were then updated by error backpropagation. The loss was minimized via standard mini-batch gradient descent with the Adam optimizer [5] and a learning rate of \(2e^{-4}\).
3 Experimental Results
We provide now a qualitative and quantitative analysis of the performance of the developed No-Reference Quality Metric (NRQM) for retinal vessel segmentation.
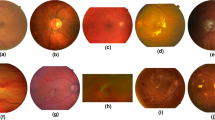
The four worse and best automatic segmentations extracted from the Messidor dataset with the technique of [9], sorted according to the proposed quality score.
3.1 Qualitative Evaluation
In a first stage, the proposed NRQM is evaluated for the task of detecting when a deficient segmentation is produced by an automatic retinal vessel extraction scheme. The Messidor-1 dataset [2], which contains 1200 images for which no manually-delineated vessel maps are available, is employed. In order to automatically produce segmentations, we apply a U-Net architecture [9]. To train it, we use the DRIVE dataset [11], which has 40 retinal images with vessel ground-truth. The model is trained on 20 images, achieving an area under the Receiver-Operator Curve (ROC) of \(97,5\%\) on the remaining images, in line with current methods.
The U-Net model produces grayscale images, referred to as soft segmentations. Each pixel contains its vessel likelihood. In order to generate a binary segmentation that can be used afterwards, e.g. to measure biomarkers related to the retinal vasculature, a binarizing threshold needs to be selected. The only feasible approach when such a system is to be deployed in a clinical scenario is to derive this threshold from the ROC curve as the one that optimizes a certain performance metric. The threshold maximizing the Youden index was selected, for which an accuracy of \(95.67\%\) was achieved on the DRIVE test set.
After selecting an optimal threshold, 1200 soft segmentations are generated from the Messidor dataset, and binarized with it. A score is computed for every segmentation, and the segmentations are sorted in descending order relative to it. Figure 4 displays the three best and worst segmentations according to the proposed NRQM. It is important to stress that the identification of these deficient segmentations was performed on a dataset without any reference ground-truth.
3.2 Quantitative Experimental Evaluation
For a quantitative evaluation of our NRQM, we selected the popular COSFIRE unsupervised vessel segmentation technique [1]. To remove any bias in our comparisons, we consider an independent test set, DRiDB [8], composed of 50 retinal images. Our evaluation of the proposed NRQM is twofold. In both cases, vessel segmentation performance will be assessed in terms of F1-score and Matthews Correlation Coefficient (MCC), which are often used within the vessel segmentation literature [1, 15] to gain evaluation insight due to the skewed-classes setting.
We compared two binarization strategies. First, ROC analysis was performed on DRIVE soft segmentations produced by COSFIRE to obtain optimal thresholds maximizing F1 score and MCC respectively. These thresholds were used to produce binary segmentations for every DRiDB image. Second, we thresholded DRiDB images at all possible values, and for each image we selected the threshold that led to a binary segmentation maximizing our NRQM. Finally, we performed statistical analysis to compare F1-scores and MCC obtained with the two approaches. Data normality was assessed using the D’Agostino-Pearson test [4]. In all cases, data were not normally distributed. Thus, we performed a nonparametric test to assess whether their population mean ranks differed using the Wilcoxon signed-rank test [13]. F1-scores and MCC of segmentations obtained using our NRQM were statistically significantly higher than those from the other compared approach, with a median difference of 0.03 for both performance metrics.
In a second stage, we investigated whether our NRQM was statistically correlated with the F1-score and MCC metrics. We calculated the F1-score and MCC for 256 uniformly distributed values of the NRQM. Again, we found that data were not normally distributed, thus we performed a nonparametric test to measure the rank correlation between the different metrics using the Spearman’s r test [10]. The proposed NRQM was very strongly positively correlated with F1-score (\(r=0.92\)) and strongly positively correlated with MCC (\(r=0.68\)).
4 Conclusions and Future Work
A no-reference metric for assessing the quality of retinal vessel maps has been introduced. Experimental results demonstrate that this score can capture the nature of deficient segmentations generated by retinal vessel segmentation algorithms, correlating well with full-reference metrics when ground-truth is available.
The approach presented here follows a general idea of modeling the degradation instead of a segmentation goal. If faithful synthetically degraded segmentations can be produced, and a meaningful similarity metric can be defined, a similar model can theoretically be trained to predict the similarity between these degraded examples and the source images. This is independent of the problem at hand, and could be explored on applications beyond retinal vessel tree segmentations.
References
Azzopardi, G., Strisciuglio, N., Vento, M., Petkov, N.: Trainable COSFIRE filters for vessel delineation with application to retinal images. Med. Image Anal. 19(1), 46–57 (2015)
Decenciére, E., et al.: Feedback on a publicly distributed image database: the messidor database. Image Anal. Stereol. 33(3), 231–234 (2014)
Gegundez-Arias, M.E., Aquino, A., Bravo, J.M., Marin, D.: A function for quality evaluation of retinal vessel segmentations. IEEE Trans. Med. Imaging 31(2), 231–239 (2012)
Ghasemi, A., Zahediasl, S.: Normality tests for statistical analysis: a guide for non-statisticians. Int. J. Endocrinol. Metab. 10(2), 486 (2012)
Kingma, D.P., Ba, J.: Adam: A Method for Stochastic Optimization. arXiv:1412.6980 [cs], December 2014
Melbourne, A., Hawkes, D., Atkinson, D.: Image registration using uncertainty coefficients. In: 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 951–954, June 2009
Pluim, J.P.W., Maintz, J.B.A., Viergever, M.A.: Mutual-information-based registration of medical images: a survey. IEEE Trans. Med. Imaging 22(8), 986–1004 (2003)
Prentašić, P., et al.: Diabetic retinopathy image database(DRiDB): a new database for diabetic retinopathy screening programs research. In: 2013 8th International Symposium on Image and Signal Processing and Analysis (ISPA), pp. 711–716, September 2013
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Spearman, C.: The proof and measurement of association between two things. Am. J. Psychol. 15(1), 72–101 (1904)
Staal, J., Abramoff, M.D., Niemeijer, M., Viergever, M.A.: Ginneken, B.v.: Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 23(4), 501–509 (2004)
Valindria, V.V., et al.: Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans. Med. Imaging 36(8), 1597–1606 (2017)
Wilcoxon, F.: Individual comparisons by ranking methods. Biom. Bull. 1(6), 80–83 (1945)
Yan, Z., Yang, X., Cheng, K.T.T.: A skeletal similarity metric for quality evaluation of retinal vessel segmentation. IEEE Trans. Med. Imaging PP(99), 1 (2017)
Zhang, J., et al.: Robust retinal vessel segmentation via locally adaptive derivative frames in orientation scores. IEEE Trans. Med. Imaging 35(12), 2631–2644 (2016)
Acknowledgments
This work is financed by the ERDF – European Regional Development Fund through the Operational Programme for Competitiveness and Internationalisation - COMPETE 2020 Programme, and by National Funds through the FCT - Fundação para a Ciência e a Tecnologia within project CMUP-ERI/TIC/0028/2014. Teresa Araújo is funded by the FCT grant contract SFRH/BD/122365/2016. The Titan Xp used for this research was donated by the NVIDIA Corporation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Galdran, A., Costa, P., Bria, A., Araújo, T., Mendonça, A.M., Campilho, A. (2018). A No-Reference Quality Metric for Retinal Vessel Tree Segmentation. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)