Abstract
Computed tomography (CT) is critical for various clinical applications, e.g., radiation treatment planning and also PET attenuation correction in MRI/PET scanner. However, CT exposes radiation during acquisition, which may cause side effects to patients. Compared to CT, magnetic resonance imaging (MRI) is much safer and does not involve radiations. Therefore, recently researchers are greatly motivated to estimate CT image from its corresponding MR image of the same subject for the case of radiation planning. In this paper, we propose a data-driven approach to address this challenging problem. Specifically, we train a fully convolutional network (FCN) to generate CT given the MR image. To better model the nonlinear mapping from MRI to CT and produce more realistic images, we propose to use the adversarial training strategy to train the FCN. Moreover, we propose an image-gradient-difference based loss function to alleviate the blurriness of the generated CT. We further apply Auto-Context Model (ACM) to implement a context-aware generative adversarial network. Experimental results show that our method is accurate and robust for predicting CT images from MR images, and also outperforms three state-of-the-art methods under comparison.
D. Nie and R. Trullo contributed equally to this work.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
CT imaging is widely used for both diagnostic and therapeutic purposes in various clinical applications. In the cancer radiation therapy, CT image provides Hounsfield units, which are essential for dose calculation in treatment planning. Besides, CT image is also of great importance for attenuation correction of positron emission tomography (PET) in the popular PET-CT scanner [7]. However, patients are exposed to radiation during CT imaging, which can damage normal body cells and further increase potential risks of cancer. It is reported that 0.4% of cancers were due to CT scanning performed in the past, and this rate will increase to as high as 1.5% to 2% in the future. Therefore, the use of CT scan should be done with great caution. MRI, on the other hand, is a safe imaging protocol which also provides more anatomical details than CT for diagnostic purpose, but unfortunately cannot be used for either dose calculation or attenuation correction. To reduce unnecessary imaging dose for patients, it is clinically desired to estimate CT images from MR images in many applications.
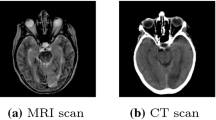
It is technically difficult to directly estimate CT image from MR image. As shown in Fig. 1, CT and MR images have very different appearances. MR images contain much richer texture information than CT images. Therefore, it is challenging to directly estimate a mapping from MRI to CT. Recently, many researches focus on estimating one modality image from another modality image, e.g., estimating CT image using MRI data. Several methods have been proposed to address this challenge. For example, Berker et al. [1] proposed to treat this problem as a segmentation task where MR images are segmented into different tissue classes and then assign each class with a known attenuation property. This method highly depends on the segmentation accuracy and always needs manual work to get final accurate results. Atlas-based methods have also been used in the literature. In [2], the authors propose to register an atlas (with the attenuation map) to the new subject’s MR image and then warp the corresponding attenuation map of the atlas to the new MR image as its estimated attenuation map. However, this kind of methods is highly dependent on the registration accuracy. On the other hand, learning-based methods learn a non-linear mapping model from MRI to CT image for alleviating the previous drawbacks. For instance, Jog et al. [6] learned nonlinear regression using random forest to improve MRI resolution. Tri et al. [5] presented an approach to predict CT image from MRI using structured random forest. Such methods often have to first represent the input MR image by features and then map them to output the CT image. Thus, the performances of these methods are bounded to the quality of the extracted features as well as how well they can represent the natural properties of the MR image.
Moreover, deep learning becomes very popular in computer vision and medical imaging fields, achieving the state-of-the-art results in both fields without the need of hand-crafted features [3, 9, 10, 12, 14]. In the particular case of image generation, Dong et al. [3] proposed to use Convolutional Neural Networks (CNN) for single image super-resolution, and Li et al. [9] applied similar deep learning models to estimate the missing PET image from the MR image of the same subject. CNNs tend to neglect neighborhood information in the predicted output image. Recently, Fully Convolutional Networks (FCN), which are a variation of the conventional CNN, have been utilized for image segmentation and synthesis so that structure information can be preserved [10, 12].
Typically, L2 distance is used as the loss function to train the previous learning based methods (e.g., random forest, CNN and FCN) for image synthesis, but it tends to produce blurry results in the output images [11]. Minimizing this distance is equivalent to maximizing the PSNR; however, as pointed out in [8], a high PSNR does not necessarily provides a perceptually better result. To address the above mentioned drawbacks, in this paper, we propose to learn the non-linear mapping from MRI to CT images through a 3D FCN. To overcome the limitations of classical L2 reconstruction, we utilize an adversarial strategy to train the FCN, which can thus enforce the generated images to be more realistic. We further propose an image-gradient-difference based loss function to alleviate the blurriness issue. Specifically, this 3D FCN is used as the generator in a generative adversarial framework, where an adversarial loss term from a discriminator network is used in addition to the conventional reconstruction error with the objective of producing more realistic CT data. The network is trained in a patch-to-patch manner, which restricts its view to the patch itself and thus cannot provide long-range information. To address this issue, we further use Auto-Context Model (ACM) where each stage is trained using the proposed framework to make it context-aware. The proposed method is evaluated on two real CT/MR datasets. Experimental results demonstrate that our method can effectively predict CT image from MR image, and also outperforms three state-of-the-art methods under comparison.
2 Methods
To address the above mentioned problems and challenges, we propose a generative adversarial network by using FCN to form the generator. First, we propose a basic 3D FCN structure to estimate the CT from MR images. Note that we use 3D operations to better model the 3D spatial information and thus could solve the discontinuity problem across slices when using the 2D CNN. Second, we utilize the adversarial training strategy [4] for the designed network, where an additional discriminator network is used to urge the generator’s output to look like the real CT as much as possible. We add an image gradient difference term to the loss function of the generator, with the goal of retaining the sharpness of the generated CT, and finally, we employ the Auto-Context Model to iteratively refine the output of the generator. At the testing stage, an input MR image is first partitioned into overlapping patches, and, for each patch, the generator is used to predict the corresponding CT patch. Then, all predicted CT patches are merged into a single CT image by averaging the intensities at overlapping CT regions. In the following paragraphs, we will describe in detail the framework used in the MRI-to-CT prediction.
2.1 Proposed Supervised Generative Adversarial Networks (GAN)
Generative Adversarial Networks (GAN) have achieved state-of-the-art results in the field of image generation producing very realistic images in an unsupervised setting [4, 13]. Inspired by the work in [4, 11], we propose a supervised GAN framework as shown in Fig. 2 to synthesize medical images. Our network includes a generator for estimating the CT and a discriminator for distinguishing the real CT from the generated CT. GANs work by training two different networks: a generator network G, and a discriminator network D. G is typically a FCN which generates images and D is a CNN which estimates the probability that an input image x is drawn from the distribution of real images; that is, it can classify an input image as real or synthetic. Both networks are trained simultaneously with D trying to correctly discriminate between real and synthetic data, while G trying to produce realistic images that will confuse D. Specifically, we minimize the binary cross entropy (bce) between the decisions of D and the correct label (real or synthetic), while the network G is trying to minimize the binary cross entropy between the decision done by D and the wrong label for the generated images, in addition to the traditional reconstruction error. In this way, D is trying to distinguish between real CT data, and the CT data generated by G. At the same time, G is trying to produce more realistic CT images such that D gets completely confused and cannot perform better than chance.
Concretely, the loss function for D can be defined as:
where X is the input MR image, Y is the corresponding CT image, and G(X) is the estimated image by the generator. \(L_{bce}\) represents the binary cross entropy.
In the case of G, as mentioned above, we use a loss function that includes an adversarial term and a reconstruction error with L2 distance. We further propose to add a gradient difference loss (gdl) as an additional term in order to deal with the inherently blurry predictions obtained from the L2 term. It is defined as:
where Y is the ground-truth CT image, and \(\hat{Y}\) is the estimated CT by the generator network. This loss function tries to minimize the difference of the magnitudes of the gradients between the ground-truth CT image and the estimated CT image. In this way, the estimated CT image will try to keep the zones with strong gradients (i.e., edges) for an effective compensation of the L2 reconstruction term. This can be approximated as finite difference during the implementation. Finally, the total loss used for training the generator G can be defined as the weighted sum of all the terms as shown in Eq. 3.
The training is performed in an alternating fashion. First, D is updated by taking a mini-batch of real CT data and a mini-batch of generated CT data (the output of G). Then, G is updated by using another mini-batch of samples including MRI and their corresponding CT. In Fig. 2, we also show the architecture of our generator network G which has the constraints mentioned above, where the numbers indicate the filter sizes. This network takes as input an MR image, and tries to generate the corresponding CT image. It has 8 stages containing convolutions, Batch Normalization and ReLU operations with number of filters 32, 32, 32, 64, 64, 64, 32, 32, respectively. The last layer only includes 1 convolutional filter, and its output is considered as the estimated CT. Regarding the architecture, we avoid the use of pooling layers since they will reduce the spatial resolution of feature maps. The Discriminator is a typical CNN architecture including three stages of convolutions+Batch Normalization+ReLU+Max Pooling, followed by one convolutional layer and three fully connected layers, where the first two use ReLU as activation function, and the last one uses sigmoid (whose output represents the likelihood that the input data is drawn from the distribution of real CT). The filter size is \(5\times 5\times 5\), the numbers of filters are 32, 64, 128 and 256 for the convolutional layers, and the numbers of output nodes in the fully connected layers are 512, 128 and 1.
2.2 Auto-Context Model (ACM) for Refinement
Since our work is patch-based, the context information available for each training sample is limited inside of the patch. This affects the modeling capacity of our network. One way to enlarge the context during the training is by using the ACM which is commonly used in the task of semantic segmentation and has been shown to be very effective [15]. In this work, we show that the ACM can also be applied successfully to the regression tasks. In particular, we adopt the ACM to iteratively refine the generated results, making our GAN context-aware. Specifically, we iteratively train several GANs that take as input the MRI patches and estimate the corresponding CT patches. These patches are then concatenated as a second channel with the MRI patches, which are used as the input for training of the next GAN.
3 Experiments and Results
We use two datasets to test our proposed methods:
-
The brain dataset was acquired from 16 subjects with both MRI and CT scans in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (see www.adni-info.org for details). The MR images were acquired using a Siemens Triotim scanner, with voxel size \(1.2\times 1.2\times 1~\text {mm}^3\), TE 2.95 ms, TR 2300 ms, and flip angle \(9^\circ \). The CT images, with voxel size \(0.59\times 0.59\times 3~\text {mm}^3\), were acquired on a Siemens Somatom scanner. A typical example of preprocessed CT and MR images is given in Fig. 1.
-
Our pelvic dataset consists of 22 subjects, each with MR and CT images. The spacings of CT and MR images are \(1.172\times 1.172\times 1~\text {mm}^3\) and \(1\times 1\times 1~\text {mm}^3\), respectively. In the training stage, CT image is manually aligned to MR image to build the voxel-level correspondence. After alignment, CT and MR images of the same patient have the same image size and spacing. Since only pelvic regions are concerned, we further crop the aligned CT and MR images to reduce the computational burden. Finally, each preprocessed image has a size of \(153\times 193\times 50\) and a spacing of \(1\times 1\times 1~\text {mm}^3\).
We extracted randomly MRI patches of size \(32\times 32 \times 32\), along with their corresponding CT of size \(16\times 16 \times 16\), using the same center points, as the paired training samples. The networks were trained using the Adam optimizer with a learning rate of \(10^{-6}\), \(\beta _1=0.5\) as suggested in [13], and mini-batch size of 10. The generator was trained using \(\lambda _1=0.5, \lambda _2=\lambda _3=1\). The code is implemented using the TensorFlow library, and we use a 4-TITAN X cluster to train our model.
To demonstrate the advantage of the proposed method in terms of prediction accuracy, we compare it with three widely-used approaches: (1) atlas-based method [16], (2) sparse representation based method, and (3) structured random forest with auto-context model [5]. We used our own implementation of the first two methods, while for the third method (structured random forest) we just show the results reported in [5]. All experiments are done in a leave-one-out fashion. The evaluation metric is the mean absolute error (MAE) and the peak signal-to-noise ratio (PSNR).
Impact of Proposed GAN Model: To show the contribution of the proposed GAN model, we conduct comparison experiments between the traditional FCN (i.e., just the generator shown in Fig. 2) and the proposed GAN model. The PSNR values are 24.7 and 25.9 for the traditional FCN and the proposed approach, respectively. These results do not include the adoption of ACM. We visualize results in Fig. 3, where the leftmost image is the input MRI and the rightmost image is the ground-truth CT. We can clearly see that the generated data using the GAN approach has less artifacts than the traditional FCN by estimating an image that is closer to the desired output quantitatively and qualitatively.
Experimental Results for Both Datasets: Considering the trade-off between the performance and the training time, we choose 2 iterations for ACM in our experiments on both datasets [15]. To qualitatively compare the estimated CT by different methods, we visualize the generated CT with the ground-truth CT in Fig. 4 (left side). We can see that the proposed algorithm can better preserve the continuity, coalition and smoothness in the prediction results, since it uses image gradient difference constraints in the image patch as discussed in Sect. 2.1. Furthermore, the generated CT looks closer to the real CT compared to all other methods. We argue that this is due to the adversarial training strategy which constrains the generated images to be very similar to the real ones, so that even a complex discriminator cannot perform better than chance.
We also quantitatively compare the predicted results in Table 1 in terms of PSNR and MAE. Our proposed method outperforms all other competing methods in both metrics, which further demonstrates the advantage of our framework.
The prediction results on the pelvic dataset by the above same methods are shown in Fig. 4 (right side). It can be seen that our result is consistent with the ground-truth CT. The quantitative results based on the same two metrics are also shown in Table 2. Quantitative results in Table 2 indicates that our method outperforms other competing methods in terms of both MAE and PSNR. Specifically, our method gives an average PSNR of 34.1, which is considerably higher than the average PSNR of 32.1 obtained by the state-of-the-art SRF+ method.
4 Conclusions
We have proposed a supervised 3D GAN model for estimating CT from MRI. Moreover, a special loss function (i.e., image gradient difference loss) is proposed to alleviate the blurry issue of the generated CT. Furthermore, the ACM strategy is adopted to make the GAN context-aware. The experiments demonstrate that our proposed method can significantly outperform three state-of-the-art methods. Note that, although we consider only the task of predicting CT from MRI, our proposed model can also be applied to other related tasks in medical application such as image super-resolution and image denoising.
References
Berker, Y., et al.: MRI-based attenuation correction for hybrid PET/MRI systems: a 4-class tissue segmentation technique using a combined ultrashort-echo-time/dixon mri sequence. J. Nucl. Med. 53(5), 796–804 (2012)
Catana, C., et al.: Toward implementing an MRI-based pet attenuation-correction method for neurologic studies on the mr-pet brain prototype. J. Nucl. Med. 51(9), 1431–1438 (2010)
Dong, C., et al.: Image super-resolution using deep convolutional networks. IEEE TPAMI 38(2), 295–307 (2016)
Goodfellow, I., et al.: Generative adversarial nets. In: NIPS, pp. 2672–2680 (2014)
Huynh, T., et al.: Estimating CT image from mri data using structured random forest and auto-context model. IEEE TMI 35(1), 174–183 (2016)
Jog, A., et al.: Improving magnetic resonance resolution with supervised learning. In: ISBI, pp. 987–990. IEEE (2014)
Kinahan, P.E., et al.: Attenuation correction for a combined 3D PET/CT scanner. Med. Phys. 25(10), 2046–2053 (1998)
Ledig. C., et al.: Photo-realistic single image super-resolution using a generative adversarial network. CoRR, abs/1609.04802 (2016)
Li, R., Zhang, W., Suk, H.-I., Wang, L., Li, J., Shen, D., Ji, S.: Deep learning based imaging data completion for improved brain disease diagnosis. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8675, pp. 305–312. Springer, Cham (2014). doi:10.1007/978-3-319-10443-0_39
Long, J., et al.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on CVPR, pp. 3431–3440 (2015)
Mathieu, M., Couprie, C., LeCun, Y.: Deep multi-scale video prediction beyond mean square error. arXiv preprint arXiv:1511.05440 (2015)
Nie, D., et al.: Fully convolutional networks for multi-modality isointense infant brain image segmentation. In: ISBI, pp. 1342–1345. IEEE (2016)
Radford, A., Metz, L., Chintala, S.: Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:1511.06434 (2015)
Shen, D., Wu, G., Suk, H.I.: Deep learning in medical image analysis. Ann. Rev. Biomed. Eng. (2017)
Zhuowen, T., Bai, X.: Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE TPAMI 32(10), 1744–1757 (2010)
Vercauteren, T., et al.: Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 45(1), S61–S72 (2009)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Nie, D. et al. (2017). Medical Image Synthesis with Context-Aware Generative Adversarial Networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10435. Springer, Cham. https://doi.org/10.1007/978-3-319-66179-7_48
Download citation
DOI: https://doi.org/10.1007/978-3-319-66179-7_48
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66178-0
Online ISBN: 978-3-319-66179-7
eBook Packages: Computer ScienceComputer Science (R0)