Abstract
Breast cancer (BC) is the most common malignancy in women worldwide. Different risk factors are associated with the disease which is classified into several intrinsic subtypes according to expression of hormone receptors (estrogen and progesterone) and human epidermal growth factor receptor 2 (HER2). The advent of high throughput techniques has allowed the molecular classification of BC and development of diagnostic/prognostic tools to predict disease outcome and progression. However, most molecular classifications and diagnostic/prognostic tools were developed using samples from European/Caucasian women. Hispanic/Latino describes an admixed group of individuals with different fractions of European, African, and Indigenous American ancestries. We determined the frequency of the different intrinsic subtypes of BC in Colombia and their association with genetic ancestry and found that luminal B subtype was the most prevalent in Colombian women with BC. We found that African ancestry was associated with more aggressive forms of BC. We then used next-generation sequencing techniques to determine the underlying markers of luminal B subtype of BC in Colombia. We found 67 genes differentially expressed between luminal A and luminal B subtypes, and of those, six were common between patients with high European/low Indigenous American ancestries. Interestingly, three genes (ERBB2, MIEN1, and GRB7) form a genomic “block” in chromosome 17 from which ERBB2 and GRB7 are co-expressed. The expression of ERBB2 was independent of HER-2 labeling. Our results suggest a major role of ethnicity in the modulation of these genes in BC in Hispanic/Latina women and may be the basis for future larger studies to investigate the role of ethnicity in BC susceptibility in minority groups.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Breast cancer is the most common cancer in women worldwide and is the second leading cause of cancer death among women in the United States [1]. Although it is the most incident cancer at the global level, its incidence and mortality rates vary among the population groups in the United States [2]. African American (AA) and Hispanic/Latina (H/L) women have a lower incidence of breast cancer (125.5 per 100,000 and 91.9 per 100,000, respectively) compared to non-Hispanic White (NHW) women (128.7 per 100,000) [2]. Mortality rates for breast cancer are higher in AA women (29.5 per 100,000) and lower in H/L women (14.2 per 100,000) when compared to NHW women (20.8 per 100,000) [2].
Breast Cancer Intrinsic Subtypes
Breast cancer is a complex and heterogeneous disease in terms of histology, therapeutic response, metastatic patterns, and outcomes [3]. In 2000, Perou et al. published the first classification of breast cancer into intrinsic subtypes based on data from gene expression microarrays [4]. They used complementary DNA (cDNA) microarrays to analyze breast cancer tissue from 65 surgical specimens of human breast tumors from 42 different patients. These samples were collected at Stanford University California, or at the Haukeland University Hospital in Bergen, Norway, so presumably included women of mostly European descent. Gene expression analysis separated intrinsic subtypes of breast cancer into two main groups based on the expression of estrogen receptor (ER). Within these two groups, four intrinsic subtypes were identified: luminal A and luminal B subtypes, which are positive for the ER expression (ER+); and HER2-enriched and basal-like, both ER negative (ER−) [5, 6]. One year later, Sorlie et al. [7] analyzed the clinical implications of this new classification of breast cancer [8] and reported differences in breast cancer outcomes between the intrinsic subtypes [7].
Genetic Ancestry and Breast Cancer Characteristics
Each Latin American country shows variations in the proportions of European, African, and Native American ancestries, and countries such as Mexico, Peru, and Bolivia are predominantly indigenous while Argentina and Uruguay are predominantly European [9]; there are differences not only in genetic ancestry but also in lifestyles and exposures of breast cancer [10].
Increasing evidence shows that breast cancer characteristics differ according to genetic ancestry. European ancestry in H/L women has been associated with an increased risk for breast cancer in women from the San Francisco Bay area [11], and this finding was replicated in women from Mexico [12]. Also, Fejerman et al. [11] did not find any associations between genetic ancestry and tumor characteristics such as hormone receptor status. Al-Alem et al. [13], however, studied the association between genetic ancestry and breast cancer characteristics in a group of 656 women (255 NHW, 277 AA, and 124 H/L) from the “Breast Cancer Care in Chicago” study and found that higher European ancestry was protective for later stage at diagnosis in H/L women (OR 0.70, 95% CI 0.54–0.92) and that Indigenous American ancestry (IAA) was associated with later stage at diagnosis (OR 1.36, 95% CI 1.04–1.79). The lack of concordance between the last two studies can be explained by the variation in the case populations. The study by Fejerman et al. [12] included women from the San Francisco Bay area, presumably of Mexican origin while the Al-Alem et al. study [13] included a larger proportion of women from the Caribbean. This highlights and reinforces the idea that results found in one Hispanic population cannot be generalized for all Latinos and that there is a need to include genetic ancestry in the studies [13].
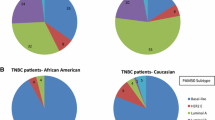
Growing evidence suggests that differences in gene expression profiles of breast cancer can be influenced by the genetic architecture of the individual’s genome [14,15,16,17]. These studies have compared gene expression profiles from NHW and AA women with breast cancer in an effort to explain health disparities in the tumor biology context. Martin et al. [16] compared breast tumors from 18 AA and 17 NHW women, and over the 400 differentially expressed genes, they found two genes that could distinguish between the two population groups, CRYBB2 and PSPHL. Similar findings were reported by Field et al. [14] who analyzed 52 matched patients (26 AA and 26 NHW) and compared the gene expression profile between the two population groups. They found 22 differentially expressed genes, including CRYBB2 and PSPHL. Stewart et al. [17], using data from The Cancer Genome Atlas (TCGA) compared gene expression profiles of 574 NHW and 53 AA patients and found 674 differentially expressed genes. Among those, resistin (RETN), a gene associated with obesity and diabetes [18], was the most changed. CRYBB2 was also found to be overexpressed in AA patients [17]. Grunda et al. [15] analyzed the expression of 84 genes involved in breast cancer prognosis associated with therapy, estrogen signaling, and tumor aggressiveness in 11 AA and 11 NHW patients and identified 20 genes that participate in regulatory processes such as G1/S transition, cell adhesion, and estrogen pathway targets. The results suggest that there may be some differences in the gene expression profile as a consequence of ancestry. None of these studies included H/L women in the analysis. More importantly, ancestry was assessed by self-identification, and genetic ancestry was not analyzed.
There is one study from Chavez-MacGregor et al. [19] that investigated the differences in gene and protein expression within each molecular subtype as a consequence of ancestry. They analyzed a group of 376 women belonging to different racial groups (AA, NHW, H/L, and others) and did not find differences in gene and protein expression between racial/ethnic groups. As they discuss in the paper, to perform a more accurate analysis in an admixed population, it is important to analyze their genetic ancestry, as it could lead to misclassification of the population. Even though the latter work did not find ancestry-modulated genes in breast cancer, more studies are needed to answer that question.
Few studies have explored genetic ancestry to assess gene expression differences. Huo et al. [20] analyzed genetic ancestry in 930 patients with breast cancer who were grouped into the categories genomic black (≥50% African ancestry) or genomic white (≥90% European ancestry). After adjusting for intrinsic subtypes, they found 142 differentially expressed genes, with LOC90784 and CRYBB2 being the top two most differentially expressed. This result is consistent with previous studies where genetic ancestry was not analyzed [14, 16, 17]. This finding can be explained by the fact that the genetic ancestry in patients who self-identify as NHW and AA is more uniform, with dominant ancestries being European and African, respectively [20].
Our recent work identified luminal B as the most prevalent subtype in Colombian women with breast cancer [21]. In the context of genetic ancestry, we analyzed gene expression profiles of 42 Colombian women with breast cancer (21 luminal A and 21 luminal B) based on 80 ancestry-informative markers (AIMs) [22]. The patients were categorized according to luminal subtype and to the proportion of European or Native American ancestry. Differential expression analysis was performed according to intrinsic subtype and by ancestry category. We found five genes potentially modulated by genetic ancestry: ERBB2, GRB7, GSDMB, MIEN1, and ONECUT2. Further studies are needed to explore the prognostic value of this finding and to replicate it in other Latin-American patients.
Other studies have analyzed gene expression profile in H/L women without determining genetic ancestry. DNA repair capacity (DRC) has been previously described as a breast cancer risk factor [23, 24]. DRC can be measured by the host-cell reactivation (HCR) assay that quantifies the capacity of a lymphocyte to repair exogenous DNA [25, 26]. Ramos et al. [27] reported that a low DRC is a breast cancer risk factor in H/L. They compared the DRC in 33 breast cancer patients and 47 healthy controls from Puerto Rico and found that for every 1% decrease in the DRC, there was a 22% increase in breast cancer risk. Matta et al. in 2012 [28] analyzed the DRC from 824 women (285 breast cancer patients and 539 controls) and also found that the DRC was lower in breast cancer patients. One year later, the same group [29] performed microarrays to analyze the expression level of DNA repair genes in women with breast cancer from Puerto Rico compared to controls to explore how DNA repair was dysregulated in breast cancer. They found 21 genes differentially expressed between breast cancer patients and controls: CHEK2, EME1 (MMS4L), ERCC3 (XPB), FANCM, H2AFX (H2AX), HMGB1, HUS1, MBD4, NEIL3, PCNA, RAD1, RAD23B, RAD51, RAD54B, RDM1 (RAD52B), SHFM1 (DSS1), TP1, UBE2N (UBC13), and XRCC5 (Ku80). Moreover, they analyzed DRC using the HCR test and found three genes positively associated with the DRC level, RAD51, FANCB, and FANCA. This study is important because the use of inhibitors of DNA repair pathways can interfere with the ability of the cells to survive DNA damage induced by chemotherapeutic agents [27, 30, 31]. The results in Matta et al. [29] and Ramos et al. [27] provide evidence regarding dysregulation of DNA repair capacity at the gene expression level in H/L women with breast cancer. Similar results have been reported in NHW women [32, 33].
Analysis of gene expression profiling has also been used to develop gene signatures to estimate recurrence risk and to better select patients who will benefit from chemotherapy [34,35,36,37]. These signatures have been developed and validated in samples of NHW women and have been used in H/L patients with the assumption that the molecular profile would be similar. Kalinsky et al. [38] compared the proliferation index of ER(+)/HER2(−) early stage tumors based on the Oncotype DX gene expression signature in H/L and NHW women and found that tumors from H/L women showed higher proliferation scores than tumors from NHW women.
Conclusions
Hispanic/Latinas are underrepresented in breast cancer studies and usually are analyzed as a whole group. However, the genetic make-up of H/L women may create a bias in genetic association studies and generate false-positive or false-negative associations. It is thus advisable to properly classify genetic ancestry in admixed populations, like Hispanic/Latinos, to better understand the real contribution of genetics to disease susceptibility and to provide this ethnic group the benefits of recent treatment advances.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–48.
Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52.
Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7(5):e1000279.
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–502.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74.
Geisler S, Lonning PE, Aas T, Johnsen H, Fluge O, Haugen DF, et al. Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res. 2001;61(6):2505–12.
Sans M. Admixture studies in Latin America: from the 20th to the 21st century. Hum Biol. 2000;72(1):155–77.
Serrano-Gomez SJ, Sanabria-Salas MC, Garay J, Baddoo MC, Hernandez-Suarez G, Mejia JC, et al. Ancestry as a potential modifier of gene expression in breast tumors from Colombian women. PLoS One. 2017;12(8):e0183179.
Fejerman L, John EM, Huntsman S, Beckman K, Choudhry S, Perez-Stable E, et al. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res. 2008;68(23):9723–8.
Fejerman L, Romieu I, John EM, Lazcano-Ponce E, Huntsman S, Beckman KB, et al. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol Biomark Prev. 2010;19(4):1074–82.
Al-Alem U, Rauscher G, Shah E, Batai K, Mahmoud A, Beisner E, et al. Association of genetic ancestry with breast cancer in ethnically diverse women from Chicago. PLoS One. 2014;9(11):e112916.
Field LA, Love B, Deyarmin B, Hooke JA, Shriver CD, Ellsworth RE. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer. 2012;118(5):1334–44.
Grunda JM, Steg AD, He Q, Steciuk MR, Byan-Parker S, Johnson MR, et al. Differential expression of breast cancer-associated genes between stage- and age-matched tumor specimens from African- and Caucasian-American women diagnosed with breast cancer. BMC Res Notes. 2012;5:248.
Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531.
Stewart PA, Luks J, Roycik MD, Sang QX, Zhang J. Differentially expressed transcripts and dysregulated signaling pathways and networks in African American breast cancer. PLoS One. 2013;8(12):e82460.
Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001;409(6818):307–12.
Chavez-Macgregor M, Liu S, De Melo-Gagliato D, Chen H, Do KA, Pusztai L, et al. Differences in gene and protein expression and the effects of race/ethnicity on breast cancer subtypes. Cancer Epidemiol Biomark Prev. 2014;23(2):316–23.
Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of breast cancer molecular features and survival by African and European ancestry in the cancer genome atlas. JAMA Oncol. 2017;3(12):1654–62.
Serrano-Gomez SJ, Sanabria-Salas MC, Hernandez-Suarez G, Garcia O, Silva C, Romero A, et al. High prevalence of luminal B breast cancer intrinsic subtype in Colombian women. Carcinogenesis. 2016;37(7):669–76.
Serrano-Gomez SJ, Sanabria-Salas MC, Garay J, Baddoo MC, Hernandez-Suarez G, Mejia JC, Garcia O, et al. Ancestry as a potential modifier of gene expression in breast tumors from Colombian women. PLoS One. 2017;12:e0183179.
Helzlsouer KJ, Harris EL, Parshad R, Perry HR, Price FM, Sanford KK. DNA repair proficiency: potential susceptibility factor for breast cancer. J Natl Cancer Inst. 1996;88(11):754–5.
Parshad R, Price FM, Bohr VA, Cowans KH, Zujewski JA, Sanford KK. Deficient DNA repair capacity, a predisposing factor in breast cancer. Br J Cancer. 1996;74(1):1–5.
Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 1991;51(21):5786–93.
Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, Kraemer KH, et al. Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res. 2002;509(1–2):165–74.
Ramos JM, Ruiz A, Colen R, Lopez ID, Grossman L, Matta JL. DNA repair and breast carcinoma susceptibility in women. Cancer. 2004;100(7):1352–7.
Matta J, Echenique M, Negron E, Morales L, Vargas W, Gaetan FS, et al. The association of DNA repair with breast cancer risk in women. A comparative observational study. BMC Cancer. 2012;12:490.
Matta J, Morales L, Dutil J, Bayona M, Alvarez C, Suarez E. Differential expression of DNA repair genes in Hispanic women with breast cancer. Mol Cancer Biol. 2013;1(1):54.
Javle M, Curtin NJ. The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer. 2011;105(8):1114–22.
Lord CJ, Ashworth A. Bringing DNA repair in tumors into focus. Clin Cancer Res. 2009;15(10):3241–3.
Shi Q, Wang LE, Bondy ML, Brewster A, Singletary SE, Wei Q. Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis. 2004;25(9):1695–700.
Xiong P, Bondy ML, Li D, Shen H, Wang LE, Singletary SE, et al. Sensitivity to benzo(a)pyrene diol-epoxide associated with risk of breast cancer in young women and modulation by glutathione S-transferase polymorphisms: a case-control study. Cancer Res. 2001;61(23):8465–9.
Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–92.
Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31(22):2783–90.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
Kalinsky K, Lim EA, Andreopoulou E, Desai AM, Jin Z, Tu Y, et al. Increased expression of tumor proliferation genes in Hispanic women with early-stage breast cancer. Cancer Investig. 2014;32(9):439–44.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Serrano-Gómez, S.J. et al. (2020). Molecular Profiles of Breast Cancer in Hispanic/Latina. In: Ramirez, A., Trapido, E. (eds) Advancing the Science of Cancer in Latinos. Springer, Cham. https://doi.org/10.1007/978-3-030-29286-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-29286-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-29285-0
Online ISBN: 978-3-030-29286-7
eBook Packages: MedicineMedicine (R0)