Abstract
Large biometric systems, e.g. the Indian AADHAAR project, regularly perform millions of identification and/or de-duplication queries every day, thus yielding an immense computational workload. Dealing with this challenge by merely upscaling the hardware resources is often insufficient, as it quickly reaches limits in terms of purchase and operational costs. Therefore, it is additionally important for the underlying systems software to implement lookup strategies with efficient algorithms and data structures. Due to certain properties of biometric data (i.e. fuzziness), the typical workload reduction methods, such as traditional indexing, are unsuitable; consequently, new and specifically tailored approaches must be developed for biometric systems. While this is a somewhat mature research field for several biometric characteristics (e.g. fingerprint and iris), much fewer works exist for vascular characteristics. In this chapter, a survey of the current state of the art in vascular identification is presented, followed by introducing a vein indexing method based on proven concepts adapted from other biometric characteristics (specifically spectral minutiae representation and Bloom filter-based indexing). Subsequently, a benchmark in an open-set identification scenario is performed and evaluated. The discussion focuses on biometric performance, computational workload, and facilitating parallel, SIMD and GPU computation.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Spectral Minutiae Representation (SMR)
- Bloom filter
- Identification
- Biometric identification
- Efficient Identification
- Binary search tree
- Column Principal Component Analysis (CPCA)
- CPCA-Tree
- Maximum curvature
- Indexing
1 Introduction
One of many catalysts for the rapid market value increase of biometrics is government-driven, large-scale biometric deployments. Most prominent examples include the Indian AADHAAR project [20], which aims to enrol the entire Indian population of 1.3 billion individuals and—at the time of writing—has already enrolled over 1.2 billion subjects, as well as several immigration programmes like the UAE or the European VIS- and EES-based border control. The operation of such large-scale deployments yields immense computational load in or duplicate enrolment checks, where—in the worst case—the whole database has to be searched to make a decision. Upscaling the hardware in terms of computing power quickly reaches certain limits in terms of, e.g. hardware costs, power consumption or simply practicability. Therefore, the underlying system’s software needs to implement efficient strategies to reduce its computational load. Traditional indexing or classification solutions (e.g. [21, 37]) are ill-suited: the fuzziness of the biometric data does not allow for naïvehashing or equality comparison methods. A good read for further understanding the problem with traditional approaches is found in [17]. This matter is the key motivation and the main focus of this chapter.
One emerging biometric characteristic that steadily increases its market shareFootnote 1 and popularity is the vascular (blood vessels) pattern in several human body parts. The wrist, back of hand and finger vessels hold the most interest since they are intuitive to capture for users and feature several advantageous properties, whereby back of hand and wrist vessels are less prone to displacement due to stretching or bending the hand. Many accurate (in terms of biometric performance) approaches and algorithms for vascular pattern recognition have emerged over time (i.e. [7, 23, 39]). However, most of them employ slow and complex algorithms, inefficient comparison methods and store their templates in an incompatible format for most template protection schemes. In other words, they generate a very high computational workload for the system’s hardware. While several biometric characteristics such as fingerprint [9] and iris [15] are already covered by workload reduction research, it is only a nascent field of research for the vascular characteristics. This chapter addresses the palm vein characteristic with a focus on the biometric identification scenario and methods for reducing the associated computational workload.
1.1 Organisation
This chapter is organised as follows:
-
Section 9.1.3 outlines requirements and considerations for the selection of the algorithms used later in this chapter.
-
Section 9.2 outlines four key computational workload reduction concepts with an algorithm proposal for each concept.
-
In Sect. 9.3, an overview of the conducted experiments using the presented concepts and algorithms is provided.
-
Subsequently, Sect. 9.4 lists and discusses the results obtained from the experiments.
-
Finally, Sect. 9.5 concludes this chapter with a summary.
1.2 Workload Reduction in Vein Identification Systems
While computational cost is not a pressing issue for biometric systems in verification mode (one-to-one comparisons), high computational costs generate several concerns in large-scale biometric systems operated in identification (one-to-many search and comparison) mode. Aside from the naïveapproach of exhaustively searching the whole database for a mated template resulting in high response times and therefore lowering usability, frustrating users and administrators, and thus lowering acceptance, another issue is presented by Daugman [12]. Accordingly, it is demonstrated that the probability of having at least one False-Positive Identification (FPI)—the False-Positive Identification Rate (FPIR)—in an identification scenario to be computed using the following formulaFootnote 2: \(\text {FPIR} = (1-(1-\text {FMR})^{N})\). Even for systems with very low FMR, this relationship is extremely demanding as the number of enrolled subjects (\(N\)) increases. Without a reduction of the penetration rate (number of template comparisons during retrieval), large biometric systems quickly reach a point where they will not behave like expected: the system could fail to identify the correct user or—even worse—allow access to an unauthorised individual. While this is less of an issue for very small biometric systems, larger systems need to reduce the number of template comparisons in an identification or Duplicate Enrolment Check (DEC) scenario to tackle the computational workload and false-positive occurrences.
Therefore, it is strongly recommended to employ a strategy to reduce the number of necessary template comparisons (computational workload reduction) for all, not only vein, modalities. As already mentioned in Sect. 9.1, computational workload reduction for vein modalities remains an insufficiently researched topic and— at the time of writing—no workload reduction approaches directly target vascular biometric systems. However, certain feature representations used in fingerprint-based biometric systems may also be applicable to vein-based systems, and hence facilitating the usage of existing concepts for computational workload reduction, as well as development of new methods. Since the vascular pattern can also be presented by minutiae (further called vein minutiae) which show almost identical characteristics compared to fingerprint minutiae, several workload reduction methods targeting minutiae-based fingerprint approaches might be usable after adaption to the more fuzzy vein minutiae.
1.3 Concept Focus
To utilise the maximum potential of the system’s hardware, all of the methods and algorithms presented in this chapter are carefully selected by the authors upon the following requirements:
-
1.
The lookup process has to be implementable in a multi-threaded manner, without creating much computational overheads (in order to manage the threads and their communication).
-
2.
The comparison algorithm has to be computable in parallel without stalling processing cores during computation.
For requirement 1, the lookup algorithm has to be separable into multiple instances, each working on a different distinct subset of the enrolment database.
In order to understand requirement 2, a brief excurse in parallel computing is needed (for a more comprehensive overview, the reader is referred to [8]). Parallel computation (in the sense of SIMD: Single Instruction, Multiple Data) is not as trivial as multi-threading where one process spawns multiple threads that are run on one or multiple CPU cores. There are multiple requirements for an algorithm to be computable in parallel, of which the two most important are as follows:
-
1.
No race conditions must occur between multiple cores.
-
2.
Multiple cores need to have the same instructions at the same time in their pipeline.
Therefore, the comparison algorithm should not rely on if-branches or jumps and the shared memory (if any) must be read-only. This results in another requirement: the feature vectors should be fixed length across all queries and templates to avoid waiting for processing templates of different sizes. However, while fixed-length template comparisons are not automatically more efficient to compute, they offer various other benefits. For example, comparisons in systems utilising fixed-length templates can usually be better optimised and implemented as simple and fast binary operations (e.g. XOR, see, for example, [16]). Furthermore, most binarisation and template protection approaches also rely on fixed-length vectors (e.g. see [22]).
Fulfilling these requirements allows for an efficient usage of SIMD instructions on modern CPUs and general-purpose GPUs (GPGPUs), hence utilising the maximum potential of the system’s hardware.
Therefore, the Spectral Minutia Representation (SMR) [35] was chosen as data representation in this chapter. Compared to shape- or graph-based approaches—like the Vascular Biometric Graph Comparison earlier introduced in this book—it fulfils all requirements: the templates using this floating-point based and fixed-length data representation can be compared by a simple image-correlation method, merely using multiplications, divisions and additions. Further, the SMR is very robust towards translations and rotations can be compensated fast. The SMR can further be binarised, which replaces the image-correlation comparison method with a simple XOR operation comparison method and thus fully allows for utilising the maximum potential of the system’s hardware. Thus, it is also compatible with various template protection approaches which rely on fixed-length binary representations. The computational efficiency of the binary SMR comparison is the main reason for the selection of the SMR as data representation. Other methods like the maximum curvature (see [24]) or Gabor filters (e.g. [38]) offer binary representations too and are less expensive in terms of computational costs while extracting the biometric features in the designated data representation. However, both the maximum curvature and the Gabor filter template comparisons are—benchmarked against the binary SMR template comparison—rather complex and expensive in terms of computational cost. Facing the high number of template comparisons needed for an identification or a duplicate enrolment check in large-scale biometric databases, the computational cost of a single SMR feature extraction is negligible with respect to the aggregate computational costs of the template comparisons. Therefore, in large-scale identification scenarios, it is more feasible to employ a computationally expensive feature-extraction algorithm with a computationally efficient comparator. Furthermore, the SMR is applicable to other modalities that can be represented by minutiae. This includes most vascular biometrics, fingerprints and palm prints. Therefore, the same method can be used for those modalities and facilitate feature-level information fusion. In particular, in this chapter, the presented system was also applied successfully for the fingerprint modality.
2 Workload Reduction Concepts
Section 9.1.2 covered the motivation behind the reduction of template comparisons in a biometric system. The same section also covered the motivation to reduce the complexity of template comparisons, namely, to achieve shorter template comparison times, thus additionally reducing the computational workload and shorten transaction times. The following sections propose components to reduce the number of necessary template comparisons and reduce the complexity of a single template comparison for a highly efficient biometric identification system. Later in the chapter, the proposed system is comprehensively evaluated.
2.1 Efficient Data Representation
Key for rapid comparisons are data representations that allow for non-complex template comparisons. Comparison subsystems and data storage subsystems can use raw minutiae (location) vectors or the vascular pattern skeletal representation as biometric templates. However, this introduces several problems, starting with privacy concerns in terms of storing the raw biometric features and ending with computational drawbacks (at least in parallel computing and the usage of CPU intrinsics) due to variable-sized feature vector sizes, whereby even probes of the same subject differ in their number of features (minutiae points). A post-processing stage can convert the raw feature vector to a fixed feature size representation that should not be reversible to the raw representation.
Inspired by the Fourier–Mellin transform [10] used to obtain a translation, rotation and scaling-invariant descriptor of an image, the SMR [18, 33] transforms a variable-sized minutiae feature vector in a fixed-length translation, and implicit rotation- and scaling-invariant spectral domain. In order to prevent the resampling and interpolation introduced by the Fourier transform and the polar-logarithmic mapping, the authors introduce a so-called analytical representation of the minutiae set and a so-called analytical expression of a continuous Fourier transform, which can be evaluated on polar-logarithmic coordinates. According to the authors, the SMR meets the requirements for template protection and allows faster biometric comparisons.
2.1.1 Spectral Minutiae Representation
In order to represent a minutiae in its analytical form, it has to be converted into a Dirac pulse to the spatial domain. Each Dirac pulse is described by the function \(m_i(x,y) = \omega (x-x_i,y-y_i),i=1,\ldots ,Z\) where \((x_i,y_i)\) represents the location of the i-th minutiae in the palm vein image. Now the Fourier transform of the i-th minutiae (\(m_i(x,y)\)) located at (x, y) is given by
with a sampling vector \(w_x\) for the angular direction and sampling vector \(w_y\) for the radial direction. Based on this analytical representation, the authors introduced several types of spectral representations and improvements for their initial approach. This chapter focuses on one of the initial representations, called the Spectral Minutia Location Representation (SML), since it achieved the best stability and thus the best biometric performance in previous experiments in [25]. It only uses the minutiae location information for the spectral representation:
In order to compensate small errors in the minutiae location, a Gaussian low-pass filter is introduced by the authors. Thus, the magnitude of the smoothed SML with a fixed \(\sigma \) is defined as follows:
in its analytical representation. By taking the magnitude—further denoted as absolute-valued representation—the translation-invariant spectrum is received (Fig. 9.1b).
When sampling the SML on a polar-logarithmic grid, the rotation of the minutiae becomes horizontal circular shifts. For this purpose, sampling of the continuous spectra (Eq. 9.3) is proposed by Xu and Veldhuis [33] using \(Xy=128\) (M in [33]) in the radial direction, with \(\lambda \) logarithmically distributed between \(\lambda _{min} = 0.1\) and \(\lambda _{max} = 0.6\). The angular direction \(\beta \) for SML is proposed between \(\beta = 0\) and \(\beta = \pi \) in \(Xx= 256\) (N in [33]) uniformly distributed samples. A sampling between \(\beta = 0\) and \(\beta = \pi \) is sufficient due to the symmetry of the Fourier transform for real-valued functions.
Since the SML yields spectra with different energies, depending on the number of minutiae per sample, each spectrum has to be normalised to reach zero mean and unit energy:
Throughout this chapter, statements that only apply for the Spectral Minutiae Location Representation will explicitly mention the abbreviation SML, while statements that are applicable to the Spectral Minutiae Representation in general will explicitly mention the abbreviation SMR.
2.1.2 Spectral Minutiae Representation—Feature Reduction
Sampling the spectra on a \(Xx= 256\) and \(Xy= 128\) grid yields a \(Xx\times Xy= {32,768}\) decimal-unit-sized feature vector. This large-scale feature vector introduces two drawbacks as given below:
- Storage:
-
Considering \(Xx\times Xy= 32{,}768\) double-precision float (64 bit) values, each template would take \(2{,}097{,}152\,\text {bit} = {256}\,\text {kB}\) RAM or data storage.
- Comparison Complexity:
-
Processing a \(Xx\times Xy= {32{,}768}\) sized feature vector is a large computational task and limits comparison speeds, especially with large-scale databases in biometric identification scenarios.
In order to address these issues, the same authors of the SMR approach introduced two feature reduction approaches in [36]. Both are based on well-known algorithms and are explained in the following subsections. In this chapter, the Column Principal Component Analysis (CPCA)—based on the idea of the well-known Principal Component Analysis (PCA) originally presented in [26]—is used. In summary, to receive the SMR reduced with the CPCA feature reduction (SMR-CPCA), the PCA only is applied to the columns of the SMR. The features are concentrated in the upper rows after applying the CPCA, and thus the lower rows can be removed, resulting in a \(Xx \times Xy_{CPCA}\) sized feature vector. According to [36], the achieved feature reduction is up to \(80\%\) by employing the SML reduced with the CPCA feature reduction (SML-CPCA) approach while maintaining the biometric performance of the original SML.
Since \(\sum _{i=1}^Z{exp(-\mathrm {j}(w_xx_i+w_yy_i))} \in \mathbb {C}\), thus \(\mathscr {M}(w_x,w_y) \in \mathbb {R}\), every element in X is defined as a 32 bit or 64 bit floating-point real-valued number. Comparisons or calculations (especially divisions) with single- or double-precision floating points are a relatively complex task compared to integer or binary operations. In order to address this computational complexity and comply with other template protection or indexing approaches where a binary feature vector is required, the SML (e.g. Fig. 9.2a, d) as well as the other SMR can be converted to a binary feature vector as presented in [32]. The binarisation approach yields two binary vectors: a so-called sign-bit vector and a so-called mask-bit vector:
- sign bit:
-
The sign-bit vector (Fig. 9.2b, e) contains the actual features of the SMR in a binary representation. Each bit is set according to one of the two binarisation approaches.
- mask bit:
-
Since binary representations suffer from bit flips on edges in fuzzy environments, a second vector (Fig. 9.2c, f) is introduced. This vector marks the likely-to-be-stable—called reliable—sign bits and is generated by applying a threshold (MT) to the spectrum.
The mask contained in the mask-bit vector is not applied to the sign bit; instead, it is kept as auxiliary data and applied during the comparison step. This approach equals the masking procedure in iris recognition (see [13, 14]).
2.1.3 Spectral Minutiae Representation—Comparison
The most proven performance in SMR comparison is reached with the so-called direct comparison.Footnote 3 It yields the most reliable comparison scores, while keeping a minimal computational complexity.
Let R(m, n) be the spectrum of the reference template and P(m, n) the spectrum of the probe template, both sampled on the polar-logarithmic grid and normalised. Then, the similarity-score \(E_{DM}^{(R,P)}\) is defined as
The score is thus defined by correlation, which is a common approach in image processing.
For comparing two binary SMRs or SMR-CPCAs, a different approach is introduced in [32], which is also used in the iris modality [13, 14].
After converting R(m, n) and P(m, n) into their individual mask bit and sign bit (see previous Sect. 9.2.1.2), yielding \(\{maskR,signR\}\) and \(\{maskP,signP\}\), the Fractional Hamming Distance (FHD) can be applied on those binary representations.
The inclusion of masks in the Hamming Distance masks out any fragile (likely-to-flip) bits and only compares the parts of the sign-bit vector where the mask-bit vectors overlap. Therefore, only the reliable areas are compared. This typically improves the recognition performance.
2.1.4 Spectral Minutiae Representation—Template Protection Properties
It is not possible to revert the spectral minutiae representation back to their initial minutiae input [33], so the irreversibility requirement of the ISO/IEC 24745 [28] standard is fulfilled. However, the spectral minutiae representation itself does not fulfil the unlinkability and renewability requirements. This issue can be tackled, e.g. with permutations of columns with application-specific keys. Depending on which templates are used in the training set of the CPCA feature reduction, a partial renewability and unlinkability (see [28]) can also be achieved, as explained in [25].
2.1.5 Spectral Minutiae Representation—Embedding Minutiae Reliability Data
It is possible that a feature-extraction pipeline may generate falsely extracted minutiae (a.k.a a spurious minutiae). Some pipelines are able to determine a genuine certainty for each minutiae, which describes the certainty that the extracted reference point is a genuine minutiae and not a spurious minutiae. When this minutiae reliability (\(q_M\), ranging 1–\(100\%\)Footnote 4) is known, the Dirac pulse (Eq. 9.1) of each minutiae can be weighted linearly (\(w_i\), ranging 0.01–1.0, corresponding to \(q_M\)) to its reliability:
Stronger reliability corresponds with a higher weight \(w_i\) for minutiae \(m_i(x,y,q_M)\). This approach is further called Quality Data-Enhanced Spectral Minutia Location Representation (QSML) throughout this chapter.
2.1.6 Spectral Minutiae Representation—Conclusions
The SML is a promising, flexible and highly efficient data representation that allows for fast comparisons using simple floating-point arithmetic in its real- or absolute-valued form. Even faster comparisons are achieved using only bit comparisons in its binary form with apparently no impairment in biometric performance. It is also possible to embed quality information. Furthermore, the SML is adaptable to template protection method which is a requirement of the ISO/IEC 24745 standard. This fixed-length representation can be compressed up to \(X{x} = 256\) and \({X}y_{CPCA} \approx 24\) bits, whereby every template is sized only 0.75 kB resulting in a 750 MB database with 1,000,000 enrolled templates.
2.2 Serial Combination of SMR
In the previous section, the SMR variant SML was introduced. As already mentioned, the SML can be represented as a real- or absolute-valued vector of its complex feature vector. Experiments in previous work (see [25]) have shown that both representations show different results in terms of comparison scores when applied on fuzzy vein minutiae. We found that for the
- absolute-valued:
-
SML, the corresponding template is less often the template with the highest score. However, on the other hand, we observed a much lower pre-selection error for rank 10 shortlists when using the absolute-valued representation compared to the
- real-valued:
-
SML, which more often fails to return a high score for the corresponding template among many templates. However, among just a few templates, the real-valued SML finds the correct template more reliably with a better score distribution than the absolute-valued SML.
The discussion of this behaviour is beyond scope of this chapter. However, this behaviour can effectively be used as an advantage by the proposed biometric system. Instead of using either absolute- or real-valued SML, both variants are incorporated: the absolute-valued representation is used during the identification lookup process to find a rank-1 to rank-10 shortlist, whereas the real-valued representation is then used to verify the rank-1 shortlist or find the correct reference template among the rank-n shortlist.
The usage of both representations does not increase the computational workload when creating the templates over the level of working with the absolute-valued representation alone since the real-valued representation is a by-product of calculating the absolute-valued representation. However, the storage requirements are doubled. Furthermore, in the shortlist, the comparison costs of the real-valued representation are also added.
2.3 Indexing Methods
In Sect. 9.2.1, an efficient data representation to effectively reduce the computational costs and time spent for template comparisons is presented. Despite the efficient data representation, the system is still subject to the challenges introduced in Sect. 9.1.2. In this section, two methods necessary to reduce the number of template comparisons are presented.
2.3.1 Bloom Filter
Following the conversion of the SML templates into their binary representation, the enrolled templates are organised into tree-based search structures by adapting the methods of [27] and [15].
-
1.
The binary SML templates are evenly split into J equally sized blocks of adjustable height and width (\({H}\times {W}\)). Subsequently, a simple transformation function is applied to the blocks column-wise, whereby each column (\(c_1,\ldots ,c_{{W}}\)) is mapped to its corresponding decimal integer value.
-
2.
For each block, an empty (i.e. all bits set to 0) Bloom filter (b) of length \(2^{{H}}\) is created and the indices corresponding to the decimal column values are set to 1 (i.e. \(\mathbf b \left[ \text {int}(c_j)\right] = 1\)).
-
3.
Hence, the resulting template (B) is a sequence of J such Bloom filters—\([\mathbf b _1,\ldots ,\mathbf b _J]\).
-
4.
The dissimilarity (DS) between two Bloom filter-based templates (denoted B and \(\mathbf B ^{\prime }\)) can be efficiently computed, as shown in Eq. 9.8, where \({|}{\cdot }{|}\) represents the population count, i.e. Hamming weight. Implementations of Eq. 9.8 can utilise intrinsic CPU operations and are trivially parallelisable, thus fulfilling the requirements stated in Sect. 9.1.3.
$$\begin{aligned} DS(\mathbf B ,\mathbf B ^{\prime }) = \frac{1}{J}\sum \limits ^{J}_{j=0}\frac{|\mathbf b _j\oplus \mathbf{b }^{\prime }_j|}{|\mathbf b _j|+|\mathbf{b }^{\prime }_j|}. \end{aligned}$$(9.8)
The Bloom filter-based templates are—to a certain degree—rotation invariant. This is because \({H}\) columns are contained within a block and hence mapped to the same Bloom filter in the sequence, which means that contrary to the raw SML, no fine alignment compensation (normally achieved via circular shifts of the template along the horizontal axis) is needed during the template comparison stage. Furthermore, the data representation is sparse, which is a crucial property for the indexing steps described below:
-
1.
The list of \(N\) enrolled templates is (approximately evenly) split and assigned to \({T}\) trees. This step is needed (for any sizeable \(N\) values) to maintain the sparseness of the data representation.
-
2.
Each node of a tree (containing
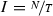 templates) is constructed through a union of templates, which corresponds to the binary OR applied to the individual Bloom filters in the sequence. The tree root is constructed from all templates assigned to the respective trees (i.e. \(\bigcup _{i=1}^{I}B_i\)), while the children at subsequent levels are created each from half of the templates of their parent node (e.g. at first level—the children of the root node—
templates) is constructed through a union of templates, which corresponds to the binary OR applied to the individual Bloom filters in the sequence. The tree root is constructed from all templates assigned to the respective trees (i.e. \(\bigcup _{i=1}^{I}B_i\)), while the children at subsequent levels are created each from half of the templates of their parent node (e.g. at first level—the children of the root node— and \(\bigcup _{i=\frac{I}{2}+1}^{I}B_i\)).
and \(\bigcup _{i=\frac{I}{2}+1}^{I}B_i\)). -
3.
The templates (\(\mathbf B _1, \ldots , \mathbf B _i\)) are inserted as tree leaves.
After constructing the trees, the retrieval can be performed as shown below:
-
1.
A small number of the most promising trees (\( t \)) out of \({T}\) constructed trees can be pre-selected (denoted \(\frac{{ t }}{{{T}}}\)) based on comparison scores between the probe and root nodes.
-
2.
The chosen trees are successively checked until the first candidate identity is found or all the pre-selected trees have been visited. Note that for the genuine transactions, thanks to the pre-selection step, the trees most likely to contain the sought identity are visited first. A tree is traversed by—at each level—computing the comparison score between its nodes and the probe, and choosing the path with the best score. Once a leaf is reached, a final comparison and check against a decision threshold takes place. The tree traversal idea is based on the representation sparseness: as long as—at each level—the relation \({DS}_{genuine}\ll {DS}_{impostor}\) generally holds true, the genuine probes will be able to traverse the tree using the correct path to reach a matching leaf template.
The complexity of a single lookup is \(O\left( {T}+ t * (2 * log_2 I)\right) \). As it is sufficient to pre-select only a small fraction of the constructed trees, i.e. \( t \ll {T}\), the lookup workload remains low, while arbitrarily many enrollees can be accommodated by constructing additional trees. For reference, Fig. 9.3 shows the indexing and retrieval in a single tree. If multiple trees are constructed, the search is trivially parallelisable by simultaneously traversing many trees at once.
2.3.2 CPCA-Tree
The second approach—called an SMR-CPCA binary search tree (CPCA-Tree)—follows the same tree construction and traversal strategy as the Bloom filter-Tree introduced in the previous section. However, instead of using a Bloom filter or another template transformation approach, the CPCA-Tree stores binary SML-CPCA templates directly. The CPCA-Tree approach has shown an advantage in terms of biometric performance over the Bloom filter-Tree in previous experiments (see [25]) when benchmarking both indexing methods with heavily degraded (i.e. very fuzzy) data since the comparison of CPCA templates does not strongly rely on stable columns like the Bloom filter. However, while the CPCA-Tree is more robust in fuzzy environments, it is to be expected that one single CPCA-Tree cannot store as many templates as one single Bloom filter-Tree: the binary SMR-CPCA features a high inter-class variance, whereby all set bits in the binary SMR-CPCA matrices are differently distributed and there are few unanimous bits. Therefore, the bits set in a binary SMR-CPCA have few bit collisions with SMR-CPCA from other subjects, respectively, from other biometric instances and when merging SMR-CPCA, the population count rises quickly, thus diminishing the descriptive value. In other words, the sparsity of upper level nodes quickly decreases to a point—typically around more than 65% of the bits set—where no correct traversal direction decisions are possible.
There are at least three approaches to store the binary SMR-CPCA templates.
- SMR-CPCA Components (SMR-CPCA-C):
-
The spectral binarisation method by Xu and Veldhuis [32] yields two binary matrices: a sign bit and a mask bit. These two matrices represent the two components of the SMR-CPCA-C representation.
- SMR-CPCA Applied (SMR-CPCA-A):
-
Instead of keeping both binary components, the mask bit is applied to the sign bit, yielding the applied bit. The applied bit matrix represents the single-component representation of SMR-CPCA-A.
- SMR-CPCA Mixed (SMR-CPCA-M):
-
Fusing both concepts by keeping the mask bit but replacing the sing bit with the applied bit.
In the experiments, the SMR-CPCA-M is used since it achieved the best biometric performance of these three representations in previous work [25]. Thus, it is required to extend the binary tree to store the applied bit and the mask bit, since both are required for the SMR-CPCA-M approach, which is commonly referred to as an auxiliary data scheme. In terms of tree construction, the applied bits are merged and the mask bits are merged upon fusing two leaves to one node.
2.4 Hardware Acceleration
Strictly speaking, the usage of hardware acceleration in the sense of multi-threaded systems, parallel systems or distinct hardware like FPGA processors is no workload reduction per se, as it does not reduce the number of template comparisons needed or reduce the size of the data. However, it is an important step to achieve an optimum efficiency of the system’s hardware and is therefore also in scope of this chapter. As already accentuated in Sect. 9.1.3, the selected approaches should be implementable in congruency with the requirements of parallel and multi-threaded systems. Our system combines two approaches (SML and indexing with binary search trees) that are evaluated for these requirements.
Implementing the binary search tree in a parallel manner is not feasible. Search trees might not be balanced or when using multiple trees, the trees differ in size. However, they are well suited for multi-threaded computation. When multiple trees were built (as would be the case in any sizeable system), each tree can be searched in one of a pool of threads. However, the SMR is perfectly suited for real parallel processing. Each element of its fixed-length feature vector can be calculated equally without any jumps or conditions. Furthermore, the calculation of one single element can be broken down to very few instructions and basic arithmetic. For example, in SSE-SIMD environments, up to four 32 bit vector elements can be calculated at a time [2] and in modern AVX-512-SIMD up to 16 32 bit vector elements at a time [1] for the real- or absolute-valued SMR. The whole calculation is also easily implementable in languages like OpenCL, which enables parallel computation on GPGPUs and other parallel systems. Next, the comparison process is also free of jumps or conditions and can also be processed in a paralleled environment where the previous statementsFootnote 5 also apply.
2.5 Fusion of Concepts
The previous sections introduced several workload reduction concepts. In fact, these concepts can be combined. This section describes the process visualised in Fig. 9.4, where all concepts are joined to one biometric system.
In terms of data processing, both the enrolment and query process are equal: after extracting the minutiae from the biometric sample, the absolute- and real-valued representations of the SML are calculated and the binary form of the absolute-valued SML is derived as introduced in Sect. 9.2.1. For the enrolment process, a binary representation (\(\mathbf {X}^b\)) of an SML template (\(\mathbf {X}\)) is now enrolled in the indexing trees and the floating-point representation (\(\mathbf {X}^f\)) is kept for each enrolled template.
Upon receiving a biometric probe that has to be identified, the binary representation is used to find a shortlist (rank-1 or rank-n) by traversing the built trees. Choosing \(n > {T}\), respective \(n > t \) is not feasible since every tree will always return the same enrolled template for the same query. Figure 9.4 is simplified to the case where \( t = n\). Subsequently, the floating-point representation of the SML query will then be compared to the real-valued SML reference templates found in the shortlist by the comparison and decision subsystem.
Accordingly, all previous concepts are fused: the binary representation—regardless of whether it is extracted from the real- or absolute-valued representation—is used to efficiently look up a small shortlist and the floating-point representation— again independent of whether it is the real or absolute valued—is used to receive a more distinct comparison score distribution. There are multiple combination possibilities, e.g. real-valued binary for enrolment and real-valued floating point for the shortlist comparison or absolute-valued binary for enrolment and real-valued floating point for the shortlist comparison. It is expected that the former yields the best biometric performance since similar experiments in [25] already revealed competitive results and it is unclear, whether the binary representation of the absolute-valued SML retains the same properties (see Sect. 9.2.2) as the floating-point SML.
3 Experiments
The following sections describe the vein data used for experiments, its preparation and a description of how the experiments to evaluate the proposed methods were conducted. This chapter merely focuses on open-set scenarios, whereby verification experiments are beyond the scope.
3.1 Experimental Setup
3.1.1 Dataset
At the time of writing, the PolyU multispectral palm-print database (PolyU) [3] is the largest publicly available vascular dataset containing Near-Infrared (NIR) palm-print images usable for (palm) vein recognition known to the authors. It comprises images of 250 subjects with 6 images per hand. The images have a predefined and stable Region of Interest (ROI). All images have a very low-quality variance and are all equally illuminated. It is not possible to link the left- and the right-hand instance of one subject by their labels and vascular pattern; therefore, every instance is treated as a single enrolment subject identifier (in short “subject”) as listed in Table 9.1. Since the PolyU dataset aims for palm-print recognition, it features a high amount of skin texture, which interferes with the vein detection and makes it a challenging dataset for the feature-extraction pipeline, which comprises the maximum curvature [24] approach with some prepended image optimisation like noise removal.
3.2 Performance Evaluation
Comparison scores are obtained in an open-set identification as follows:
-
1.
One reference template is enrolled for each subject.
-
2.
All remaining probe templates are compared against the enrolled references.
-
3.
The scores are then categorised as
- false-positive identification:
-
identification transactions by data subject not enrolled in the system, where an identifier is returned.
- false-negative identification:
-
identification transactions by users enrolled in the system in which the user’s correct identifier is not among those returned.
The dataset has been split into four groups: enrolled, genuine, impostor and training (for the CPCA feature reduction). An overview of the relevant numbers is listed in Table 9.2. In order to ease the indexing experiments, an enrolment set of \(2^n\) is preferred.
With a limited number of subjects (500), 256 enrollees offer the best compromise between largest \(2^n\)-enrolment and the number of impostor queries.
The results of the experiments are reported as a Detection Error Trade-off Curve (DET). To report the computational workload required by the different approaches, the workload metric
where \(N\) represents the number of enrolled subjects, p represents the penetration rate and \({C}\) represents the costs of one single one-to-one template comparison (i.e. number of bits that are compared), and the fraction
introduced by Drozdowski et al. [15] will be used.
In tables and text, the biometric performance is reported with the Equal Error Rate (EER). However, when evaluating the best biometric performance, the results are first ordered by the False-Negative Identification Rate (FNIR) at FPIR \( = 0.1\%\), then ordered by the EER. This is due to the nature of the EER, whereby it does not describe the biometric performance at important low FPIR, i.e. an experiment with EER \( = 5\%\) can feature an FNIR at FPIR \( = 0.1\%\) of \(20\%\) while an experiment with EER \(= 5.5\%\) can feature an FNIR at FPIR \( = 0.1\%\) of \(13\%\). In real-world scenarios, the latter example result is more relevant than the former.
3.3 Experiments Overview
The following enumeration serves as an overview of the experiments conducted in this chapter:
- Spectral Minutiae Representation:
-
The basic implementation, comparing vein probes based on minutiae with the SML in both absolute- and real-valued representation. In an identification scenario, the database is searched exhaustively, e.g. every query template (probe) is compared with every enrolled template (reference). These experiments represent the biometric performance and workload baseline for the workload reduction approaches in the following experiments.
- CPCA Feature Reduction:
-
Repetition of the above identification experiments, but with CPCA feature reduction for both binary and floating-point SML—further called SML-CPCA—to evaluate whether the biometric performance suffers from the feature reduction.
- Binary Spectral Minutiae Representation:
-
The same experiments (for both SML representations) as above are repeated with the binary representations of the SML to evaluate whether the biometric performance is degraded by this binarisation process.
- Serial Combination of SML:
-
With the baseline for all representations of the SML,Footnote 6 these experiments are used to validate the assumption that the observed advantages of both SML representations can be used to increase the biometric performance.
- Indexing Methods:
-
The binary representations of both absolute- and real-valued SML are indexed with the presented Bloom filter-Trees and CPCA-Trees approaches to evaluate whether the biometric performance is degraded by these indexing schemes.
- Fusion of Concepts:
-
Both indexing and serial combination of SML will be combined as presented in Sect. 9.2.5. This experiment evaluates whether both concepts can be combined to achieve a higher biometric performance due to the serial combination, combined with a low computational workload due to the indexing scheme.
4 Results
This section reports and comments on the results achieved by the experiments presented in the previous section.
4.1 Spectral Minutiae Representation
The SML experiments are split in multiple stages to approximate its ideal settings and tuning for fuzzy vascular data.
4.1.1 Baseline
In order to assess the results of the main experiments (indexing approaches), a baseline is needed. Figure 9.5a shows the DET curves of the introduced SML and QSML in both real- and absolute-valued sampling. It is clearly visible that the real-valued representation is much more accurate than the absolute-valued representation. Furthermore, Fig. 9.5b contains plots of the real-valued SML, QSML and Spectral Minutia Location Representation with minutiae pre-selection (PSML) thresholds of *0.1, 0.2 and 0.3.
While the authors of [31, 34] recorded good results using the absolute-valued sampling for their verification purposes, it falls far behind the real-valued sampling in identification experiments.
The selected dataset introduces some difficulties for the feature-extraction pipeline used. Recall that the PolyU dataset is a palm-print and not palm vein dataset, and therefore it includes the fuzzy skin surface, which would not be included in a designated vascular dataset. It is mainly selected because due to its size rather than quality. Various optimisation experiments were run and are reported in the following section to increase the recognition performance. Implementing a robust feature extractor is beyond the scope of this chapter.
4.1.2 Optimisation
The feature extractor (maximum curvature [24]) used is able to report quality (reliability; \(q_M\)) data in the limits of (0, 1]—where 1 represents a \(100\%\) minutiae reliability and 0 no minutiae at all—about the extracted pattern; therefore, the QSML can be used. Using this data to remove unreliable minutiae in terms of defining a \(q_M\) threshold (i.e. a minutiae reliability of at least \(20\%\)), the recognition performance can be increased as shown in Fig. 9.5b. Using \(q_M\ge 0.2\) as a threshold for the so-called PSML and quality data-enhanced Spectral Minutia Location Representation with minutiae pre-selection (PQSML) achieved the best results in the experiments.
Additionally, it is possible to reduce the SML and QSML samplings \(\lambda _{max}\) to fade out higher (more accurate) frequencies, which increases the significance of the lower, more stable (but less distinct) frequencies. Experiments showed that using \(\lambda _{max}\approx 0.45\) instead of the original \(\lambda _{max}\approx 0.6\) resulted in the best compromise between low and high frequencies. This optimisation process is further referred to as tuning.
4.1.3 CPCA
In order to investigate the impact of the CPCA compression on the recognition performance, the same procedure as for the SML and QSML is repeated using the CPCA compression.
Applying CPCA to the tuned SML and QSML results in no noticeable performance drop, as shown in Fig. 9.6. Again, using \(\lambda _{max}\approx 0.45\) instead of the original \(\lambda _{max}\approx 0.6\) resulted in the best compromise between low and high frequencies. One mentionable result of these experiments is that the tuned QSML-CPCA performs slightly better than the full-featured and tuned QSML.
4.1.4 Summary
In summary, even with a moderately reliable feature-extraction pipeline, the SML achieved acceptable results. Employing quality data in terms of minutiae reliability improved the biometric performance and an additional \(\lambda _{max}\)-tuning also improved the biometric performance (as shown in Fig. 9.7). For the following experiments, the tuned QSML-CPCA with minutiae pre-selection of \(q_M\ge 0.2\) will be used as a biometric performance baseline and will further be called PQSML-CPCA. The corresponding workload for the SMLFootnote 7 is \(W \approx 2.52\times 10^7\).
4.2 Binary Spectral Minutiae Representation
The next SML optimisation step is to binarise the SML floating-point vector. This step shrinks the feature vector by a factor of 32 and enables the usage of highly efficient binary and intrinsic CPU operations for template comparisons. Intrinsic CPU operations are also available for floating-point values. However, the binary intrinsics are favourable since they are more efficient and allow for a higher number of feature vector element comparisons with a single instruction. Practically, it is possible to binarise the full-featured SML, as well as the more compact SML-CPCA. However, it holds special interest to achieve a high biometric performance with the binarised (P)SML-CPCA or (P)QSML-CPCA to receive the smallest possible feature vector. Interestingly, the binary CPCA-reduced variants perform better than their larger counterpart, as is visible in the DET plots of Fig. 9.8. Moreover, the binary QSML-CPCA outperforms its minutiae-pre-selection counterparts. By analysing the other binary QSML-CPCA results, this result could be a coincidence. At this point, the \(256\times 128\) floats sized (PQ)SML got shrunk to a \(256\times 20\)-bit sized (PQ)SML-CPCA without exhibiting a deterioration of the biometric performance. The workload for the binary (PQ)SML-CPCAFootnote 8 is only at \(W \approx 1.31\times 10^6\). This results in \(\digamma \approx 5\%\) for the binary (PQ)SML-CPCA compared to the full-featured (PQ)SML.
4.3 Serial Combination of SMR
The serial combination of PQSML experiments was run with different settings ranging from rank-1 to rank-25. Only the PQSML was experimented with since it mostly performed better than the other representations. Using a rank-10 to rank-15 (\({\sim }5\%\)) pre-selection with the absolute-valued PQSML then comparing the real-valued PQSML templates of the generated shortlist achieved the best results as shown in Fig. 9.9. Both were sampled with the same settings, whereby only one SMR sampling is needed; recall that, the real-valued SMR is a by-product when calculating the absolute-valued SMR. However, it is questionable whether the EER decrease achieved justifies the introduced online workload \(W\approx 2.64\times 10^7\) (\(\digamma \approx 105\%\)), compared to the \(W\approx 2.52\times 10^7\) calculated for the (PQ)SML, yielded by this method if the shortlist is not generated using the efficient, binary representation.
4.4 Indexing Methods
The previous experiments demonstrated that it is possible to reduce the workload drastically without a major impairment of the biometric performance by compressing and subsequently binarising the PQSML. However, it is still necessary to exhaustively search the whole database. In this section, the results of the indexing experiments conducted to reduce the number of necessary template comparisons are reported.
4.4.1 Bloom Filter
First experiments showed a severely impaired biometric performance loss of about 15% points (Fig. 9.10) compared to the results reported in Sect. 9.4.2 when employing Bloom filter indexing. The origin of the poor performance of the applied to binary PSML-CPCA templates is the high number of bit errors when comparing two mated binary PSML-CPCA. This Bloom filter implementation strongly relies on stable columns in the J blocks of the binary vector across their height H to offer a high biometric performance. The iris naturally yields comparatively stable columns when aligned and unrolled and therefore the Bloom filter performs exemplary. However, due to the nature of the SMR— which includes various frequencies—this stability is not ensured: smaller feature-extraction inconsistencies yield much more noise in the upper frequencies of the SMR, which then result in more Bloom filter errors, mostly along the columns. A more in-depth discussion of this behaviour is given in Sect. 9.4.2 of [25]. Even at a very high MT of 0.9, the average bit error rate is 13% with an error in more than 50% of the columns, which is excessive for a reliable Bloom filter transformation that needs stable column bits.
While analysing the issue in further depth, it was found that the Bloom filter reliably looked up correct templates for genuine queries but failed to achieve a separable score distribution. Therefore, the Bloom filter indexing might not be feasible if used on its own, although it performs well in a serial combination approach. The best biometric performance was recorded at  resulting in a workloadFootnote 9 of \(W\approx 7.7\times 10^5\), achieving \(\digamma \approx 3.1\%\).
resulting in a workloadFootnote 9 of \(W\approx 7.7\times 10^5\), achieving \(\digamma \approx 3.1\%\).
4.4.2 CPCA-Tree
In its basic implementation, the CPCA-Tree surpasses the basic Bloom filter indexing in both the FNIR-to-FPIR ratio, as well as EER. It achieves a similar EER to the naïve binary QSML-CPCA and naïve QSML-CPCA. Thus, the CPCA-Tree indexing approach reaches a similar biometric performance as the naïve approaches, albeit with a much lower workloadFootnote 10 of \(W\approx 6.8\times 10^5\), which results in \(\digamma = 1.7\%\).
Therefore, if a serial combination approach is not desired because due to its complexity, the CPCA-Tree is a good compromise between complexity, workload and biometric performance.
4.5 Fusion of Concepts
As already mentioned in the experiment’s description, the fusion of concepts combines the serial combination of (PQ)SML and the indexing schemes following the scheme presented in Sect. 9.2.5. In the first run of the experiment, \(\mathbf {X}^b\) was extracted from the real-valued QSML and \(\mathbf {X}^f\) is the real-valued QSML, and out of \( t \) selected trees only one (rank-1) template was selected for the shortlist. While this did not affect the biometric performance of the CPCA-Tree indexing, the Bloom filter indexing transcends the biometric performance of the CPCA-Tree indexing approach for lower FPIR with the rank-1 serial combination scheme. However, the Bloom filter indexing could not catch up at higher FPIR rates. Using a higher pre-selection rank for the Bloom filter indexing scheme did not result in a higher biometric performance.
In these experiments, the pre-selection rank is set equal to the number of searched trees \( t \). Upon first glance, the results of the higher pre-selection rank experiments for the CPCA-Tree indexing do not deviate much compared to the rank-1 experiments, whereby only the EER is slightly lower. Note the number of searched trees \( t \), with a higher rank, a comparable biometric performance is achieved by traversing lesser trees. This is an important property for scaling in large-scale databases. For medium-scale databases, the overhead introduced by the additional floating-point comparisons when comparing the query with the templates in the shortlist would void the workload reduction achieved by the reduction of traversed trees. Furthermore, the experiments using a real-valued pre-selection/real-valued decision achieved a higher biometric performance than the absolute-valued pre-selection/real-valued decision and the absolute-valued pre-selection/absolute-valued decision. Therefore, the statement of Sect. 9.2.2 that the absolute-valued SML is better suited for lookup but the real-valued SML yields more distinctive comparison scores does not apply on the binary representation of the absolute-valued (PQ)SML. The recorded workloads in this experiment are consolidated in Table 9.3 and the DET curves are shown in Fig. 9.11.
4.6 Discussion
Most results have already been discussed in previous sections. Finally, at least three properties for a new biometric deployment have to be considered when choosing one of the presented approaches: scalability, complexity and biometric performance. If a system simple to implement is desired, the CPCA-Tree indexing is recommended, given that it is easy to implement and it achieved biometric performance comparable with the contestants. Conversely, if the implementation complexity is less an issue, scalability and biometric performance have to be considered. In terms of scalability, the rank-n serial combination is the recommended approach, whereby it achieved a biometric performance comparable with that of the other approaches at the smallest number of traversed trees (smallest computational workload). Regarding the biometric performance, the rank-1 serial combination real/real indexing scheme achieved the best results. Table 9.4 summarises the rating for all best performing configurations of each approach from best (\(++\)) to worst (\(--\)) with gradations of good (\(+\)), neutral (o) and bad (−).
To deterministically benchmark the different indexing methods and configurations, the Euclidean distance between the baseline operation point (\(B_{{EER}} = 5.5\%\), \(B_{\digamma }=1\%\)) and the best performing configuration of each approach—as shown in Eq. 9.11—can be used.
The smaller the \(\Delta (TP,{\digamma })\) for an approach, the closer that its point of operation is to the baseline operation point, whereby smaller is more preferable. Choosing the baseline operation point (\(\digamma =1\%\), \({EER}=5.5\%\)) instead of the optimal operation point (\({EER}=0\%\), \(\digamma \approx 0\%\)) moves the emphasis of the distance to the performance of the indexing schemes rather than to the performance of the baseline system.
The data of Table 9.5 is visualised as scatterplot in Fig. 9.12. Note that the naïvePQSML system is not plotted since \(\digamma =100\%\) would render the y-axis scaling of the plot impractical.
Scatterplot of Table 9.5
5 Summary
Vascular patterns are an emerging biometric modality with active research and promising avenues for further research topics. With the rising acceptance of biometric systems, increasingly large-scale biometric deployments are put into operation. The operation of such large deployments yields immense computational load. In order to maintain a good biometric performance and acceptable response times—to avoid frustrating their users—computational workload reduction methods have to be employed. While there are many recognition algorithms for vascular patterns, most of them rely on inefficient comparison methods and hardly any computational workload reduction approaches for vein data can be found.
A recently published biometric indexing approach based on Bloom filters and binary search trees for large-scale iris databases was adopted for vascular patterns. In order to further develop this indexing approach, the vascular pattern skeletal representation of the raw palm vein images was extracted and the minutiae—the endpoints and bifurcations—of the extracted vascular pattern were then transformed using a Fourier transformation based approach originally presented for the fingerprint characteristic. When transforming the floating-point representation yielded by the Fourier transformation to a binary form, it is possible to apply the Bloom filter indexing. It has been demonstrated that the Bloom filter indexing system is capable of achieving a biometric performance close to the naïvebaseline, while reducing the necessary workload by an additional \({\approx }37\%\) on top of the workload reduction achieved with the CPCA compression and binarisation. Some of the approaches used by the Bloom filter in [15] were not feasible and the fuzziness of the vascular pattern prevented a higher workload reduction without losing too much biometric performance. However, the most important approaches have been successfully applied, and thus the system appears to be scalable in terms of workload reduction, biometric performance and enrollees.
An additional, less complex, biometric indexing approach merely using a reduced form of the binary Fourier transformation representation and binary search trees has been presented. It adopts most workload reduction strategies that are used for the Bloom filter indexing approach and achieved a better biometric performance with only a slightly lower computational workload reduction (compared to a naïveimplementation using the reduced binary Fourier representation) of \(\approx 36\%\). Since the presented approach follows the same theory and implementations as the binary search trees of the Bloom filter indexing, it also appears to be scalable in terms of workload reduction, biometric performance and enrollees.
The respective advantages and disadvantages of the two indexing methods were outlined based on the results from the previous sections. It has been shown that the CPCA-Tree achieves good performance with less stable templates than the Bloom filter. However, it is to be expected that the Bloom filter will outperform the CPCA-Tree approach with more stable templates. Furthermore, the potential for computational workload reduction is much higher using the Bloom filter based method.
The overall workload is reduced to an average of 3% compared to the baseline of the naïveimplementation using the Fourier representation in both systems. All approaches used are perfectly implementable in either multi-threaded or parallel environments. The presented indexing approaches are well suited to run in multiple threads yielding hardly any overhead. Furthermore, the data representation used can efficiently be computed and compared with SIMD introduction and intrinsics, whereby both computation and comparison do not rely on jumps or conditions. Therefore, it is perfectly suited for highly parallel computation on GPGPUs or manycore CPUs, hence utilising the maximum potential of the system’s hardware.
The workload reduction approaches achieved very promising results, which were doubtless limited by the biometric performance of the base system. It is to be expected that with a higher biometric baseline performance, a higher workload reduction can be achieved: with more stable templates, a more robust indexing can be achieved, thus further reducing the workload. Several early experiments and approaches in [25] already achieved a significant biometric baseline performance gain (\(EER < 0.3\%\)), which will be used in future work. Since the base system achieved a very high biometric performance for fingerprints, the workload reduction approaches can be adopted to the fingerprint modalities and is subject to future work.
Finally, it should be noted that there is a lack of publicly available large (palm-)vein datasets (with more than 500 palms) suitable for indexing experiments. Most datasets comprise only 50–100 subjects (100–200 palms). In order to fairly and comprehensively assess the computational workload reduction and scalability of indexing methods, large-scale data is absolutely essential. As such, entities (academic, commercial and governmental alike) that possess or are capable of collecting the requisite quantities of data could share their datasets with the academic community, thereby facilitating such evaluations. Another viable option is an independent benchmark (such as, e.g. FVC Indexing [6], IREX one-to-many [4] and FRVT 1:N [5] for fingerprint, iris and face, respectively), which could also generate additional interest (and hence research) in this field from both the academic and the commercial perspective. Lastly, the generation of synthetic data (e.g. finger veins [19]) is also a possibility, albeit on its own, it cannot be used as a substitute for real large-scale data.
Notes
- 1.
- 2.
This equation ignores other error sources like failure-to-acquire (FTA).
- 3.
In [35], the comparison method is named direct matching, where matching is used as a non-ISO compliant synonym for the term comparison.
- 4.
A reliability of \(0\%\) should not be possible since no minutiae would have been detected in the first place.
- 5.
It has to be noted, that for the binary SMR, up to two rows can be processed with one AVX-512-SIMD instruction.
- 6.
Real-valued, absolute-valued, real-valued SML-CPCA, absolute-valued SML-CPCA, binary real-valued SML-CPCA and binary absolute-valued SML-CPCA.
- 7.
\(N=256, p=1, {C}=256\times 128\times 3\); Measurements on the machine running the experiments resulted in three times slower floating-point SML comparisons than binary SML bit comparisons.
- 8.
\(N=256, p=1, {C}=256\times 20\).
- 9.
\(N= 256,Xy_{CPCA}= 20, {W}= 5, {H}= 4, {C}= (2^{{H}}*\frac{Xy_{CPCA}}{{H}})*\frac{Xx}{{W}} = 4096, p = \frac{{T}+ t * (2 * log_2(\frac{N}{{T}}))}{N} \approx 0.73\).
- 10.
\(N= 256, Xy_{CPCA}= 20, {C}= Xx*Xy_{CPCA}, p = \frac{{T}+ t * (2 * log_2(\frac{N}{{T}}))}{N} \approx 0.51\).
References
Intel intrinsics guide: _mm_mul512_ps. https://software.intel.com/sites/landingpage/IntrinsicsGuide/#expand=3675,3675,3681&techs=AVX_512&text= _mm512_mul_ps
Intel intrinsics guide: _mm_mul_ps. https://software.intel.com/sites/landingpage/IntrinsicsGuide/#expand=3675,3675&techs=SSE&text=_mm_mul_ps
Polyu multispectral palmprint database. http://www.comp.polyu.edu.hk/~biometrics/MultispectralPalmprint/MSP.htm
Iris exchange (irex) (2017). https://www.nist.gov/programs-projects/iris-exchange-irex-overview
Face recognition vendor test (frvt) 1:n 2018 evaluation (2018). https://www.nist.gov/programs-projects/frvt-1n-identification
Fvc-ongoing indexing (2019). https://biolab.csr.unibo.it/fvcongoing/UI/Form/BenchmarkAreas/BenchmarkAreaFIDX.aspx
Abbas A, George LE (2014) Palm vein recognition and verification system using local average of vein direction. Int J Sci Eng Res 5(4):1026–1033
Baxter S (2011) Modern GPU—introduction to GPGPU—GPU performance. http://web.archive.org/web/20111007211333/, http://www.moderngpu.com/intro/performance.html
Cappelli R, Ferrara M, Maltoni D (2011) Fingerprint indexing based on minutia cylinder-code. IEEE Trans Pattern Anal Mach Intell 33(5):1051–1057
Casasent D, Psaltis D (1976) Position, rotation, and scale invariant optical correlation. Appl Opt 15(7):1795–1799
Credence Research: Biometrics technology market by technology (face recognition, hand geometry, voice recognition, signature recognition, iris recognition, afis, non-afis & others), end use vertical (government, military and defense, healthcare, banking and finance, consumer electronics, residential and commercial security, transportation and immigration & others)—growth, share, opportunities & competitive analysis, 2015–2022 (2016). http://www.credenceresearch.com/report/biometrics-technology-market
Daugman J (2000) Biometric decision landscapes. University of Cambridge, Computer Laboratory, Tech. rep
Daugman J (2003) The importance of being random: statistical principles of iris recognition. Pattern Recogn 36(2):279–291
Daugman J (2004) How iris recognition works. IEEE Trans Circ Syst Video Technol 14(1):21–30
Drozdowski P, Rathgeb C, Busch C (2017) Bloom filter-based search structures for indexing and retrieving iris-codes. IET Biom
Drozdowski P, Struck F, Rathgeb C, Busch C (2018) Benchmarking binarisation schemes for deep face templates. In: 2018 25th IEEE international conference on image processing (ICIP). IEEE, pp 191–195
Hao F, Daugman J, Zielinski P (2008) A fast search algorithm for a large fuzzy database. IEEE Trans Inf Forensics Secur 3(2):203–212
Hartung D, Olsen MA, Xu H, Busch C (2011) Spectral minutiae for vein pattern recognition. In: 2011 international joint conference on biometrics (IJCB). IEEE, pp 1–7
Hillerstrom F, Kumar A, Veldhuis R (2014) Generating and analyzing synthetic finger vein images. In: 2014 international conference of the biometrics special interest group (BIOSIG). IEEE, pp 1–9
India: Aadhaar issued summary. https://portal.uidai.gov.in/uidwebportal/dashboard.do
Indyk P, Motwani R (1998) Approximate nearest neighbors: towards removing the curse of dimensionality. In: Proceedings of the thirtieth annual ACM symposium on theory of computing. ACM, pp 604–613
Lim M, Teoh ABJ, Kim J (2015) Biometric feature-type transformation: making templates compatible for secret protection. IEEE Sig Process Mag 32(5):77–87
Lin S, Tai Y (2015) A combination recognition method of palmprint and palm vein based on gray surface matching. In: 2015 8th international congress on image and signal processing (CISP). IEEE, pp 567–571
Miura N, Nagasaka A, Miyatake T (2007) Extraction of finger-vein patterns using maximum curvature points in image profiles. IEICE Trans 90-D(8):1185–1194. http://dblp.uni-trier.de/db/journals/ieicet/ieicet90d.html#MiuraNM07
Mokroß BA (2017) Efficient biometric identification in large-scale palm vein databases. Master’s thesis, Hochschule Darmstadt. https://dasec.h-da.de/wp-content/uploads/2017/12/Thesis-mokross-vein_indexing.pdf
Pearson K (1901) On lines and planes of closest fit to systems of points in space. Lond Edinb Dublin Philos Mag J Sci 2(11):559–572
Rathgeb C, Baier H, Busch C, Breitinger F (2015) Towards bloom filter-based indexing of iris biometric data. In: ICB. IEEE, pp 422–429. http://dblp.uni-trier.de/db/conf/icb/icb2015.html#RathgebBBB15
Rathgeb C, Busch C (2012) Multi-biometric template protection: Issues and challenges. In: New trends and developments in biometrics. IntechOpen
Tractica LLC: Biometrics market forecasts: Global unit shipments and revenue by biometric modality, technology, use case, industry segment, and world region: 2016–2025 (2017). https://www.tractica.com/research/biometrics-market-forecasts/
Transparency Market Research: Biometrics technology (face, hand geometry, voice, signature, iris, afis, non-afis and others) market—global industry analysis size share growth trends and forecast 2013–2019 (2014). https://www.transparencymarketresearch.com/biometrics-technology-market.html
Xu H, Veldhuis RN (2009) Spectral minutiae representations of fingerprints enhanced by quality data. In: IEEE 3rd international conference on biometrics: theory, applications, and systems, 2009. BTAS’09. IEEE, pp 1–5
Xu H, Veldhuis RN (2010) Binary representations of fingerprint spectral minutiae features. In: 2010 20th international conference on pattern recognition (ICPR). IEEE, pp 1212–1216
Xu H, Veldhuis RN (2010) Spectral minutiae representations for fingerprint recognition. In: 2010 Sixth international conference on intelligent information hiding and multimedia signal processing (IIH-MSP). IEEE, pp 341–345
Xu H, Veldhuis RN, Bazen AM, Kevenaar TA, Akkermans TA, Gokberk B (2009) Fingerprint verification using spectral minutiae representations. IEEE Trans Inf Forensics Secur 4(3):397–409
Xu H, Veldhuis RN, Kevenaar TA, Akkermans AH, Bazen AM (2008) Spectral minutiae: a fixed-length representation of a minutiae set. In: IEEE computer society conference on computer vision and pattern recognition workshops, 2008. CVPRW’08. IEEE, pp 1–6
Xu H, Veldhuis RN, Kevenaar TA, Akkermans TA (2009) A fast minutiae-based fingerprint recognition system. IEEE Syst J 3(4):418–427
Zezula P, Amato G, Dohnal V, Batko M (2006) Similarity search: the metric space approach, vol 32. Springer Science & Business Media
Zhang H, Liu Z, Zhao Q, Zhang C, Fan D (2013) Finger vein recognition based on gabor filter. In: Sun C, Fang F, Zhou ZH, Yang W, Liu ZY (eds) Intelligence science and big data engineering. Springer, Berlin, Heidelberg, pp 827–834
Zhou Y, Kumar A (2011) Human identification using palm-vein images. IEEE Trans Inf Forensics Secur 6(4):1259–1274
Acknowledgements
This work was partially supported by the German Federal Ministry of Education and Research (BMBF), by the Hessen State Ministry for Higher Education, Research and the Arts (HMWK) within the Center for Research in Security and Privacy (CRISP), and the LOEWE-3 BioBiDa Project (594/18-17).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Mokroß, BA., Drozdowski, P., Rathgeb, C., Busch, C. (2020). Efficient Identification in Large-Scale Vein Recognition Systems Using Spectral Minutiae Representations. In: Uhl, A., Busch, C., Marcel, S., Veldhuis, R. (eds) Handbook of Vascular Biometrics. Advances in Computer Vision and Pattern Recognition. Springer, Cham. https://doi.org/10.1007/978-3-030-27731-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-27731-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27730-7
Online ISBN: 978-3-030-27731-4
eBook Packages: Computer ScienceComputer Science (R0)







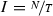 templates) is constructed through a union of templates, which corresponds to the binary OR applied to the individual Bloom filters in the sequence. The tree root is constructed from all templates assigned to the respective trees (i.e.
templates) is constructed through a union of templates, which corresponds to the binary OR applied to the individual Bloom filters in the sequence. The tree root is constructed from all templates assigned to the respective trees (i.e.  and
and 








