Abstract
An available recognition toolkit is one of the basic requirements for conducting research in finger- and hand vein recognition. Currently, there is only one comprehensive, open-source, software package available, which includes a full finger vein recognition toolkit. We present a novel, full fledged vein recognition software framework implemented in MATLAB. Our PLUS OpenVein software package contains various well-established and state-of-the-art vein enhancement, feature extraction and comparison schemes. Moreover, it contains tools to evaluate the recognition performance and provides functions to perform feature- and score-level fusion. It is fully open source and available free of charge for research and non-commercial purposes. This vein recognition framework has already been used in several papers and can be a valuable tool for other researchers working in vein recognition.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Finger vein recognition
- Hand vein recognition
- Open-source software
- Vein recognition software
- Performance evaluation
1 Introduction
Vascular pattern-based biometrics are an emerging biometric trait due to their various advantages over other biometric traits. Vascular pattern-based biometrics deal with the patterns formed by the blood vessels located inside the human body, where the patterns inside the human fingers and hands are the most widely used body parts. This is commonly denoted as finger- and hand vein recognition. In order to conduct research on any biometric trait, there are two important prerequisites: the availability of datasets for training and testing and the availability of a complete biometric recognition tool chain tailored to the specific biometric trait to be able to conduct recognition performance evaluations.
Regarding the first requirement, the availability of datasets, there are already several publicly available finger- and hand vein data sets like the UTFVP [45], the HKPU [22], the SDUMLA-HMT [52] and the FV-USM [3]. Regarding open finger vein sensors, besides the sensor presented by Ton [44] and Raghavendra et al. [36], we propose an open- source finger vein scanner design which is available free of charge for research purposes in Chap. 3 [13] of this book. This enables every researcher to capture his own data set with a reproduction of a scanner based on our design. Hence, it is easily possible to meet the first requirement by either obtaining one of the publicly available datasets or by capturing a new one.
Prior to publishing any new results in vein recognition, regardless if they are from a proposed enhancement scheme or a new feature extraction approach, the second important requirement is the implementation of a complete biometric recognition tool chain. This tool chain includes the following five major steps/modules: The first one is parsing the dataset and reading the images according to the subject, finger/hand and image ID. The second step is preprocessing in order to enhance the quality of the vein patterns. The next step is the feature extraction itself, where feature vectors or biometric templates, encoding the relevant details of the vein patterns are extracted from the preprocessed images. During the subsequent comparison step, two templates are compared against each other and a comparison score is calculated. The last step is the performance evaluation, where certain performance figures and plots describing the recognition performance of the evaluated approach are generated. These results are a vital part in assessing the performance of a new vein recognition scheme and publishing a paper without reporting recognition performance results hardly makes sense. Another important aspect is that it should be easy to include a new recognition scheme in the existing tool chain in order to evaluate its recognition performance. Of course, it is always possible to combine all the individual parts and implement this tool chain from scratch for conducting the performance evaluation. However, it is more convenient if there is a tool chain provided and only the new part has to be included in the existing tool chain. Moreover, if the tool chain provides standardised test protocols it helps in enabling a common basis for the performance evaluation and makes the results easier to compare.
For several parts of the recognition tool chain, there are a few publicly available implementations available, especially for the feature extraction and comparison step. There is only one full featured vein recognition library, which is part of the BOB library, written in Python, combining all the necessary steps, available. Even this library, which is described in the following subsection, does neither include more recent vein recognition schemes, nor a comprehensive set of vein tailored enhancement schemes. BOB provides such a tool chain where the test protocols are included and a new recognition scheme can be easily added by adhering to the specified interface. Although Python and especially some freely available Python modules provide several image processing operations, many researchers prefer MATLAB instead of other programming languages to test their new approaches. MATLAB provides many common image processing operations out of the box and is easy to use and debug.
In this chapter, we present a new full fledged, open-source, vein recognition framework, called PLUS OpenVein Toolkit which is written in MATLAB. This framework includes a full vein recognition tool chain, consisting of image reading, preprocessing, feature extraction, comparison and performance evaluation. Its design enables an easy integration of new recognition schemes and allows for flexible combinations of different preprocessing and feature extraction schemes. It supports several commonly used hand- and finger vein datasets out of the box while it is straightforward to include the support for new ones. There are 11 vein specific preprocessing as well as 13 feature extraction and comparison approaches included. The whole framework is available free of charge for research and non-commercial purposes. In addition, we would encourage every interested researcher to contribute their own work to extend and improve the whole framework.
The rest of this chapter is structured as follows: Sect. 4.2 discusses related work on publicly available vein recognition software. The software framework, its history, licensing, the directory structure as well as the contained external components are explained in Sect. 4.3. The vein recognition schemes included in our OpenVein toolkit are listed and described in Sect. 4.4. Section 4.5 gives some exemplary baseline results of the toolkit’s application on the UTFVP dataset. Section 4.6 summarises this chapter together with an outlook on future work.
2 Related Work
Bram Ton provided several finger vein recognition methods as MATLAB implementations on MATLAB Central.Footnote 1 These implementations include the following feature extraction methods: Maximum Curvature, proposed by Miura et al. [32], Repeated Line Tracking, also proposed by Miura et al. [31], Principal Curvature, proposed by Choi et al. [6] and the Wide Line Detector as proposed by Huang et al. [8]. The comparison scheme used by Miura et al. in [31, 32] is also contained. In addition, the finger region detection method proposed by Lee et al. [23] and the finger normalisation method proposed by Huang et al. [8] are included too. However, there is no surrounding framework combining all those individual parts and forming a whole vein recognition tool chain (e.g. to read all the images of a whole dataset and to evaluate the recognition performance). We included all the schemes implemented and provided by Bram Ton in our software framework. The respective links can be found in Sect. 4.3.3.
The Bob toolboxFootnote 2 [1, 2], which “is a signal-processing and machine learning toolbox originally developed by the Biometrics Security and Privacy Group, and the Research and Development Engineers at Idiap, in Switzerland.” contains a biometric vein recognition library.Footnote 3 This library is based on their open-source biometric recognition platform, using the base types and techniques provided by the bob.bio.base package.Footnote 4 The bob.bio.vein package contains several finger vein preprocessing and feature extraction/comparison schemes as well as performance evaluation tools and supports a few publicly available vein datasets. It is written in Python and uses several Python libraries like NumPiFootnote 5 and SciPi.Footnote 6 The implemented vein preprocessing methods include fixed cropping, masking and padding, histogram equalisation, finger normalisation as proposed by Huang et al. [8], finger masking as proposed by Kono [21], finger masking as proposed by Lee et al. [23] and a Watershed segmentation-based finger masking. The provided feature extraction methods consist of: an LBP based approach [30], Miura et al.’s Maximum Curvature [32] and Repeated Line Tracking [31] method, a normalised cross-correlation method as proposed by Kono [21], Choi et al.’s Principal Curvature [6] and the Wide Line Detector proposed by Huang et al. [8]. The comparison algorithms include a simple cross-correlation based method, a Hamming distance-based approach and the cross-correlation approach suggested by Miura et al. [31, 32]. The bob.bio.vein software runs automated biometric recognition experiments where a certain dataset, preprocessor, feature extractor and comparison algorithm are selected. The results can be saved and several evaluation metrics and plots can be generated. Score-level fusion is supported too. The UTFVP [45] and the Vera Fingervein database [43] are supported out of the box. A new or custom data set can be easily defined following their interface specification. Furthermore, the software package is able to perform a grid-based best parameter search and utilise parallel processing.
Our provided PLUS OpenVein Toolkit vein recognition framework is an all-in-one solution, providing a complete biometric recognition tool chain from reading the input vein images to outputting the performance figures and results. It includes 11 vein specific preprocessing and image enhancement schemes together with some more generic ones. Thirteen different feature extraction methods together with the respective comparison schemes are implemented or included, ranging from several well-established ones to more recent state-of-the-art ones like DTFPM [27] and ASAVE [51]. Moreover, several evaluation protocols are available together with some widely used performance figures and plots. It is able to process finger- as well as hand vein images. Furthermore, basic feature- and score-level fusion tools as well as image contrast and vein image quality assessment schemes are provided as well. It supports a variety of different finger- and hand vein datasets. All those individual parts are bundled in an easy and convenient to use MATLAB-based vein recognition framework, which is available free of charge. The included schemes are described in the following section.
3 PLUS OpenVein Toolkit
This subsection gives a general overview of the vein recognition framework, including its structure and main components as well as its terms of use and where to obtain it.
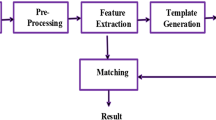
Figure 4.1 shows the design of a typcial biometric recognition system, including the biometric trait which is to be captured, the biometric capture device and the recognition tool chain. Our PLUS OpenVein Toolkit vein recognition framework resembles the software parts, i.e. the whole recognition tool chain part of the figure. The corresponding methods are shown in Fig. 4.2. The whole framework is implemented in MATLAB. At the time of writing, MATLAB version 2016a–2018b are supported. Compatibility to new releases will be reported on our website. MATLAB is widely used in academia as it enables quick and easy prototyping as well as platform independence. MATLAB itself is neither open source nor free of charge. However, many research institutions already licensed MATLAB so there will be no additional costs for them. Moreover, student licenses for MATLAB are available at a very reasonable price. Futhermore, there is the free and open source alternative to MATLAB, called GNU Octave.Footnote 7 Note that the toolkit has not been tested in Octave and is not guaranteed to run with Octave.
The vein recognition framework started as a finger vein recognition tool in the scope of a proseminar project back in 2013 which was done by Christof Kauba and Jakob Reissig. At first, only MC, RLT and WLD were contained as feature extraction methods, based on the publicly available code provided by B.T. Ton on MATLAB Central. Then a SIFT-based feature extraction method [14] and several preprocessing methods have been implemented. From there on it started to grow and include several other feature extraction and preprocessing schemes as well as convenience code for automated execution based on settings files before we finally decided to provide it to the research community as an open-source framework. The PLUS OpenVein Toolkit vein recognition framework is availableFootnote 8 free of charge for research and non-commercial purposes. It is covered under the New BSD Licence.Footnote 9 By downloading and using our framework you agree to our terms of use, especially to cite one of our papers [11, 13] if results obtained with the help of our recognition framework are published. Further usage instructions, a detailed description of the individual parts and a link to our git repository can be found on the website. The framework is still being improved and extended and we encourage every interested researcher to contribute to our open-source framework.Footnote 10 It has already been used in numerous publications (sorted by date of publication):
-
Kauba, C., Reissig, J., Uhl, A.: Pre-processing cascades and fusion in finger vein recognition (2014) [14]
-
Kauba, C., Uhl, A.: Sensor Ageing Impact on Finger-Vein Recognition (2015) [16]
-
Kauba, C., Uhl, A.: Robustness Evaluation of Hand Vein Recognition Systems (2015) [15]
-
Kauba, C.: Impact of Sensor Ageing on Finger-Image based Biometric Recognition Systems. Master’s thesis (2015) [9]
-
Kauba, C., Piciucco, E., Maiorana, E., Campisi, P., Uhl, A.: Advanced Variants of Feature Level Fusion for Finger Vein Recognition (2016) [10]
-
Kauba, C., Uhl, A.: Shedding Light on the Veins—Reflected Light or Transillumination in Hand-Vein Recognition (2018) [18]
-
Kauba, C., Uhl, A.: Robustness of Finger-Vein Recognition (2018) [17]
-
Kauba, C., Prommegger, B., Uhl, A.: Focussing the Beam—A New Laser Illumination Based Data Set Providing Insights to Finger-Vein Recognition (2018) [11]
-
Kauba, C., Prommegger, B., Uhl, A.: The Two Sides of the Finger—An Evaluation on the Recognition Performance of Dorsal versus Palmar Finger-Veins (2018) [12]
-
Prommegger, B., Kauba, C., Uhl, A.: Multi-Perspective Finger-Vein Biometrics (2018) [34]
-
Prommegger, B., Kauba, C., Uhl, A.: Longitudinal Finger Rotation—Problems and Effects in Finger-Vein Recognition (2018) [33]
-
Kirchgasser, S., Kauba, C., Uhl, A.: Towards Understanding Acquisition Conditions Influencing Finger-Vein Recognition (2019) [20]
-
Prommeger, B., Kauba, C., Uhl, A.: A Different View on the Finger—Multi-Perspective Score Level Fusion in Finger-Vein Recognition (2019) [35].
3.1 Directory Structure
Figure 4.3 shows a schematic overview of the vein recognition framework. The main file is Matcher.m, which contains most of the program logic, including the preprocessing, feature extraction, comparison execution functions. The “matcher” is actually a MATLAB object, also storing the input images, the extracted features, the comparison scores and the results. Some parts of the recognition schemes are directly implemented in Matcher.m, but most of the schemes are called as external functions, implemented in distinct .m files. These .m files are organised in the following subdirectories:
-
Automation: several scripts for batch automation, automated testing of setting files, etc.
-
EEREvaluation: functions to determine the performance figures and plots.
-
FeatureExtraction: most of the self-contained feature extraction functions, like the ones provided by Bram Ton.
-
FeatureLevelFusion: tools to perform feature-level fusion and evaluate fusion results.
-
GUI: graphical user interface related stuff.
-
Matching: different comparison functions, e.g. the Miura matcher.
-
Preprocessing: various preprocessing functions for vein images.
-
Quality Assessment: a few general image contrast metrics as well as vein specific quality metrics.
-
ScoreLevelFusion: tools and functions to perform (simple) score-level fusion.
-
Settings: contains the settings files for various datasets.
-
UtilityFunctions: several helper functions, e.g. for handling ini files, progress bar, plotting SIFT key-points.
-
vl_feat: directory where to put the vl_feat sources.
Besides these directories, there are several other directories inside the main directory, e.g. Matsuda16 and ASAVE. These recognition schemes contain feature extraction as well as comparison methods and it is not obvious how to separate these parts so we decided to put all the stuff necessary for the scheme in one directory, named according to the method. As this is neither feature extraction only, nor comparison only, the directories are located inside the main directory instead. More details on the implemented recognition and evaluation schemes can be found in Sect. 4.4.
Each step of the program execution can be called manually. To run the whole recognition tool chain at once, including reading of the vein images, preprocessing, feature extraction, comparison and performance determination, we provide the automateMatcher() function/script, which has to be called with the paths to the input images and the desired settings file, which is described in Sect. 4.3.2. During each of the steps, a progress bar with the current progress and the estimated remaining time is shown. After all the steps are finished, the results (plots and EER/FMR values) are displayed.
3.2 Settings Files
Several settings files for the supported datasets are provided in the Settings subdirectory. There is an example settings file in this directory, called settingsExample.ini, which lists all possible options (excluding all parameters of the different preprocessing, feature extraction and comparison methods) together with a short explanation. All the important parameters and options are controlled via these settings files and by automatically generating settings files, various different settings can be tested in batch processing for parameter tuning. The settings are grouped according to general settings, preprocessing settings, feature extraction settings, optional post-processing settings, comparison settings and results settings. Optionally, most of these options can be passed to automateMatcher as additional input arguments, overwriting the parameters defined in the settings file. The settings are described in more detail in the readme file of the PLUS OpenVein Toolkit.
3.3 External Dependencies
Our framework contains several external dependencies. The following ones are not included in the sources of the framework and have to be downloaded separately and then put into the respective subdirectories:
-
vl_feat: mainly used to provide a SIFT implementation, the MATLAB version can be found at [68].
-
MASI Fusion: implements the STAPLE/STAPLER and COLLATE fusion algorithms. The MATLAB version is available at [62].
-
ARIA Vessels Library: used to perform the IUWT feature extraction, a MATLAB implementation is freely available at [57].
Furthermore, several schemes and functions have not been implemented by ourselves. Instead, publicly available implementation have been used. These implementations are already contained in the framework:
-
MC, WLD, RLT vein network extraction methods: these implementations are provided by Bram Ton and are available on MATLAB Central [65] and [67].
-
PC: For principal curvature [6] another implementation of Bram Ton is used.
-
LeeRegion, HuangNormalise: these implementations are provided by Bram Ton and are available on MATLAB Central [66] and [64].
-
Retinex: a publicly available MATLAB implementation of Brian Funt, Florian Ciurea, and John McCann is utilised. It can be found at [59].
-
SUACE: The implementation provided by the original authors of [4] is used. It is available on github: [56].
-
Adaptive thresholding: an implementation by Guanglei Xiong, which is available at [69] is used.
-
Gaussian filtering: an implementation by F. van der Heijden, available as part of the Image Edge Enhancing Coherence Filter Toolbox [60] is used.
-
Frangi filtering and Hessian filtering: an implementation by D. Kroon, also available as part of the Image Edge Enhancing Coherence Filter Toolbox [60] is utilised.
-
Histogram distances: For several of the included histogram comparison distances the implementation by Boris Schauerte, available at [63] are used.
-
EER/DET plots: For determining the EER and other performance figures as well as for creating the ROC/DET plots, modified versions of the functions contained in the BioSecure framework are utilised. The original implementation can be found at: [61].
-
Ini file handling: The open source ini library for MATLAB by Primoz Cermelj is utilised, available at: [58].
For several basic image processing as well as morphological image operations, functions provided by MATLAB’s image processing toolbox are utilised. All the other vein specific preprocessing, feature extraction and comparison schemes that are included within the PLUS OpenVein Toolkit and not listed above are custom re-implementations done by the authors. Some smaller helper functions and methods are not implemented by the authors, but again publicly available implementations have been utilisied. These are not listed above, however, the information regarding the original author and the copyright notice can be found in the respective source code files. More details about each individual implementation can be found in the source code files of the respective scheme.
4 Included Vein Recognition Schemes
In this section, the reading of the input datasets, preprocessing, feature extraction, comparison and performance evaluations schemes which are implemented and/or included in our PLUS OpenVein Toolkit are listed and described, including references to their original publications. Some of those methods are custom re-implementations done by the authors of the PLUS OpenVein Toolkit while for others, publicly available implementations or the original authors’ implementations have been used if available. Section 4.3.3 lists the details about the schemes which are used from external sources and have not been implemented by ourselves.
4.1 Input File Handling/Supported Datasets
Instead of acquiring the biometric data directly from the capture device, this data is read from the file system in the form of images. The handling of the image files (getting all files in a directory, reading the images and storing them in an in-memory cell array for further use) is done in readImages.m. Parsing of the file names (subject, finger/hand and sample ID) is based on regular expressions. A new dataset can be easily added by adding a new case to the switch-case clause in readImages.m, providing a suitable regular expression for parsing the file name. Currently, the following commonly used hand- and finger vein dataset are supported by our vein recognition framework Footnote 11:
-
UTFVP [45],
-
SDUMLA-HMT [52],
-
FV-USM [3],
-
HKPU [22],
-
VERA FingerVein Spoofing [43],
-
VERA PalmVein [42],
-
PLUSVein-FV3 [12],
-
PLUSVein-3D-FV [35],
-
PROTECT Hand-/ Finger-/ and Wrist Vein [46].
4.2 Preprocessing
This subsection lists the included preprocessing schemes, grouped according to vein tailored ones at first, followed by general image processing ones. The names in brackets are the names as used in the toolkit. All of the mentioned preprocessing schemes can be combined in any order and more than once (a list of methods and parameters has to be provided in the settings file), e.g. CLAHE followed by Zhao09, followed by Zhang09, followed by CLAHE, followed by Resize. Different parameters for each preprocessing scheme can be used each time it is applied. The order and the parameters are configured in the settings file.
Finger Masking (LeeRegion)
Lee et al. [23] proposed a simple method to localise the finger region and mask out the background. In general, the finger region is brighter than the background. Thus, to detect the finger outline, the image is convolved with a custom mask containing lines of 1 and \(-1\) values (different order for upper and lower outline). Afterwards, all pixels above the detected upper boundary and below the detected lower boundary are masked out by setting their value to 0 (black).
Finger Rotation Compensation Normalisation (HuangNormalise)
Huang et al. [8] use a normalisation method based on the finger outline and the finger baseline as the first step of their finger vein recognition approach. They utilised an active contour model to detect the finger outline. Afterwards, the baseline of the finger is estimated as a straight line by calculating the midpoints between the points of the top and bottom outline of the finger using the least squares method. The angle between the finger baseline and the horizontal line is then determined and the image is transformed such that the finger baseline is horizontal. The transformation is done by the nearest sampling method. In our implementation, the LeeRegion method is used to detect the finger outline instead of the active contour model.
High-Frequency Emphasis Filtering (Zhao09)
Zhao et al. [54] utilised high-frequency emphasis filtering in combination with histogram equalisation for improving hand vein images. High-frequency Emphasis Filtering (HFEF) is applied in the frequency domain, hence at first the Fourier transform of the input vein images is computed. The HFE is computed based on an offset and a fixed multiplicative factor together with a Butterworth high-pass filter. Then the inverse Fourier transform is computed to obtain the enhanced image. Finally, a simple global histogram equalisation is applied to further improve the image contrast. Instead of this global histogram equalisation, we use CLAHE in our implementation.
Grey Level Grouping with Circular Gabor Filtering (Zhang09)
A combination of Grey Level Grouping and circular Gabor filters was proposed by Zhang and Yang [53] in the context of finger vein image enhancement. At first, they applied Grey Level Grouping to reduce illumination fluctuations and improve the contrast of the finger vein images. Then a circular Gabor filter is used to further enhance the visibility of the vein ridges in the images. In contrast to usual Gabor filters, which have a certain direction, Circular Gabor Filters (CGF) are omnidirectional. A CGF is essentially a 2D band pass filter whose passband looks like a circle and which is suitable for edge detection in arbitrary directions. Hence, vein lines in arbitrary directions are captured without distortion by this type of Gabor filter. The authors only use the real part of the Gabor filter output, thus arriving at an even-symmetric circular Gabor filter. Instead of Grey Level Grouping, we apply CLAHE again as the first step in our implementation.
Multichannel Gabor Filtering for Finger Vein Image Enhancement (Yang09)
Yang and Yang [50] proposed a finger vein enhancement scheme based on multichannel Gabor filters in different scales and orientations. The input vein image is filtered by various different Gabor filters and the outputs of the individual filters are then combined to a final enhanced vein image using a simple reconstruction rule where a pixel value at a certain position is set to the index of the Gabor filter with the lowest output value. The Gabor filters are designed in a way that their period is twice the vein width and at least the orientation of one of the filters should be orthogonal to the vein lines. Four orientations with different centre frequencies turned out to achieve the best enhancement results.
A New Algorithm for Finger Vein Image Enhancement and Segmentation (Shi12)
The vein image enhancement scheme proposed by Shi et al. [38] is based on Koschmieder’s law to reduce light scattering effects and on a bank of even Gabor filters to further enhance the visible vein patterns. At first, an anisotropic diffusion method together with an averaging filter is used to generate a smoothed finger vein image. This image in combination with the assumption that the extinction coefficient and the thickness of the skin tissue are used to solve the Koschmieder’s law equation with respect to the undistorted vein image. This image is then further enhanced by applying a bank of even-symmetric Gabor filters. The final output image is obtained by combining the responses of each single Gabor filter using a multi-scale multiplication rule to further suppress unwanted information.
Finger Vein Image Restoration Based on a Biological Optical Model (Yang12)
Instead of Koschmieder’s law, Yang et al. [49] proposed another vein enhancement scheme based on a Biological Optical Model (BOM). This model is based on the Beer–Lambert law, describing the light transport attenuation in the tissue and the non-scattered transmission map, describing the optical transmissivity of the given tissue. Solving this model from a single observed image is an ill-posed problem, hence the scattering component has to be estimated in order to solve this model. At first, the vein image is transformed to its negative version, which is beneficial for scattering illumination estimation. Then the scattering component is estimated by introducing three constraints. Afterwards, the scattering radiation is estimated based on local pixel statistics. Finally, the original finger vein image can be restored (enhanced) by computing a pixel-wise restoration based on the proposed BOM.
Finger Vein Ridge Enhancement (YangShi12)
Yang and Shi [48] suggested another finger vein enhancement scheme, again based on Koschmieder’s law and a bank of even-symmetric Gabor filters, like they did in [38]. In contrast to their previous work, they did not use a simple multi-scale multiplication rule to combine the outputs of each individual Gabor filter to the final enhanced vein image, but they only use the single output of a filter per pixel based on the reconstruction rule introduced in [50]. Afterwards, they apply the multi-scale multiplication rule of [38] together with a normalisation rule to arrive at the enhanced vein image. This should help to suppress false vein ridge information.
Intensity Variation Normalisation for Finger Vein Recognition Using Singe Scale Retinex (Retinex)
Inspired by the work of Xie et al. [47] we included a single scale Retinex implementation in our framework. We did not include the guided filter part of the paper though. The Retinex method [28] is based on the assumption that an observed image can be regarded as the multiplication of several illumination and reflectance images. Reflectance changes sharply while illumination changes only smoothly. The Retinex method decomposes the intensity image (input image) in those two images (illumination and reflectance) by trying to estimate the illumination image using a Gaussian filter and subtracting the illumination image from the intensity image in the logarithm domain. The remaining reflectance image, resembling an illumination-normalised output, is the Retinex output.
Contrast Limited Adaptive Histogram Equalisation (CLAHE)
In contrast to global histogram equalisation, Adaptive Histogram Equalisation (AHE) is a local technique, computing several histograms for distinct sections of the image. These histograms are equalised individually and then combined in order to redistribute the lightness values of the image. AHE tends to over amplify the contrast in homogeneous regions of the image as the histogram in these regions is concentrated (only a few distinct grey values occur). Contrast Limited AHE (CLAHE) [55] limits the contrast amplification, in order to reduce the problem of noise amplification. CLAHE has been successfully used as a simple means of enhancing the contrast and quality of vein images. MATLAB’s implementation of CLAHE as provided by the adapthisteq function is utilisied in our framework.
Speeded Up Adaptive Contrast Enhancement (SUACE)
The contrast enhancement method proposed by Bandara et al. [4] is, especially, tailored to enhance superficial vein images. In contrast to traditional approaches which are based on costly adaptive histogram equalisation methods, SUACE uses a fixed range for enhancing the contrast all over the image, which is shifted according to the calculated illumination at the current pixel position. Therefore, it utilises the response from low-frequency range of the infra-red input image signal, which is calculated by filtering the original image with a Gaussian filter, to adjust the boundaries for the reference dynamic range. This dynamic range is then used in a linear contrast stretching process.
Further Filtering/Image Processing Schemes
In addition to the above-mentioned, vein specific, preprocessing methods, we included several more generic image processing schemes which are able to enhance the vein image quality. These further schemes include image resize, image cropping, image rotation, median filtering, Wiener filtering, unsharp masking and Gaussian high-pass filtering. Most of these methods are based on the image processing functions provided by MATLAB.
4.3 Feature Extraction
In the following, the included feature extraction methods are described. These methods are grouped by vein based ones, i.e. outputting a binary vein image, followed by key-point based ones and general purpose ones.
Maximum Curvature (MC)
This feature extraction technique proposed by Miura et al. [32] aims to emphasise only the centre lines of the veins and is therefore insensitive to varying vein widths. The first step is the extraction of the centre positions of the veins by determining the local maximum curvature in cross-sectional profiles obtained in four directions: horizontal, vertical and the two oblique directions. The cross-sectional profile is determined based on the first and second derivates. Then each profile is classified as either being concave or convex, where only the local maxima belonging to a concave profile indicate a vein line. Afterwards, a score according to the width and curvature of the vein region is assigned to each centre position and recorded in a matrix called locus space. Due to noise or other distortions, some pixels may not have been classified correctly at the first step. Thus, the centre positions of the veins are connected using a filtering operation in all four directions taking the 8-neighbourhood of pixels into account. The final binary output image is obtained by thresholding of the locus space using the median as a threshold.
Enhanced Maximum Curvature (EMC)
EMC is an extension of Maximum Curvature by Syarif et al. [41]. In addition to the original MC approach, there is an additional image filtering/enhancement step based on Hessian Vessel filtering in between the extraction of the centre position of the veins and the filtering operation to connect the centre positions in order to extract small vein delineation that is hardly visible in the previously extracted vein patterns. Furthermore, a Histogram of Oriented Gradients (HOG) descriptor is used as feature representation instead of a simple binarisation fo the extracted curvature information as in the original Maximum Curvature. HOG captures edge and gradient structures and thus, the local gradient information of the vein features, more effectively than the simple thresholding based binarisation. In our vein recognition framework, there are two versions of EMC: one that only adds the additional enhancement step, called EMC within the framework and the other one, which is a full implementation of the method as proposed by Syarif et al., called EMC_HOG within the framework.
Wide Line Detector (WLD)
The Wide Line Detector [8] is essentially an adaptive thresholding technique (using isotropic non-linear filtering), i.e. thresholding inside a local neighbourhood region. The difference of the centre pixel to its neighbours inside a circular neighbourhood and the number of pixels inside this neighbourhood with a difference smaller than a predefined threshold are determined. This number is again thresholded to get the final binary output vein image.
Repeated Line Tracking (RLT)
As the veins appear as valleys in the cross-sectional profile of the image, RLT [31] tries to track the veins as dark lines inside the image. The tracking point is repeatedly initialised at random positions and then moved pixel by pixel along the dark line, where the depth of the valley indicates the movement direction. If no “valley” is detected a new tracking operation is started. The number of times a pixel is tracked, is recorded in a matrix. Pixels that are tracked multiple times as belonging to a line statistically have a high likelihood of belonging to a blood vessel. Binarisation using thresholding is applied to this matrix to get the binary output image.
Principal Curvature (PC)
Choi et al. [6] proposed this curvature-based feature extraction method. At first the gradient field of the image is calculated. In order to prevent the unwanted amplification of small noise components, a hard thresholding which filters out small gradients by setting their values to zero is done. Then the gradient at each pixel is normalised to a magnitude of 1 to get a normalised gradient field. This normalised gradient field is smoothed by applying a Gaussian filter. The next step is the actual principal curvature calculation. The curvatures are obtained from the Eigenvalues of the Hessian matrix at each pixel. The two Eigenvectors of the Hessian matrix represent the directions of the maximum and minimum curvature and the corresponding Eigenvalues are the principal curvatures. Only the bigger Eigenvalue which corresponds to the maximum curvature among all directions is used. The last step is a threshold-based binarisation of the principal curvature values to arrive at the binary vein output image.
Gabor Filtering (GF)
Gabor filters are inspired by the human visual system’s multichannel processing of visual information and have been widely used in biometrics. A Gabor filter is a Gaussian kernel function modulated by a sinusoidal plane wave. Kumar and Zhou [22] proposed a Gabor filter based finger vein extraction approach. Therefore, a filter bank consisting of several 2D even-symmetric Gabor filters with different orientations (in \(\frac{\pi }{k}\) steps where k is the number of orientations) is created. k feature images are extracted by filtering the vein image using the different filter kernels contained in the Gabor filter bank. The final feature image is obtained by summing up all the single feature images from the previous step and thresholding the resulting feature image. This image is then post-processing using morphological operations to remove noise and to get the final binary vein output image.
Anatomy Structure Analysis-Based Vein Extraction (ASAVE)
Yang et al. [51] proposed a new finger vein feature extraction and comparison approach based on incorporating knowledge about the anatomical structure (directionality, continuity, width variability as well as smoothness and solidness) and imaging characteristics of the underlying vein patterns. The vein pattern is extracted by their orientation map-guided curvature based on valley- or half-valley-shaped structures. This curvature is extracted using the Maximum Curvature algorithm [32]. The vein pattern is then further thinned and refined (filling, thinning, denoising and connecting) using morphological operations to make it more reliable. Furthermore, a so-called vein backbone is extracted from the input vein image by thresholding the curvature image, retaining only the most clear and reliable vein branches. This vein backbone is then utilised to align two images during comparison in order to compensate for horizontal as well as vertical displacements. Comparison itself is done using an elastic matching approach, which is able to tolerate small deformations in the two vein patterns. The similarity score is further refined by incorporating the overlap degree of the two vein backbones to arrive at the final output score.
Isotropic Undecimated Wavelet Transform (IUWT)
IUWT [40] is a special type of Wavelet transform which can be implemented in a simple way. At each iteration j, the scaling coefficients \(c_{j}\) are computed by low-pass filtering and the wavelet coefficients \(w_{j}\) by subtraction. The subsequent scaling coefficients of the transform are calculated by a convolution of the jth scale’s scaling coefficient with a filter \(h^{j}\). The subsequent wavelet coefficients are the difference between two adjacent sets of scaling coefficients. Levels 2 and 3 exhibit the best contrast for blood vessels and are thus taken into account for feature extraction. The vein features are extracted by adding the wavelet levels 2 and 3. The final features are obtained by thresholding the resulting Wavelet transform. This binarised image is then post-processed with morphological operations to remove noise.
Deformation Tolerant Feature Point Matching (DTFPM)
This key-point-based technique proposed by Matsuda et al. [27] replaces the conventional SIFT descriptor and key-point detector by vascular pattern tailored ones, taking the curvature and the vein directions into account. This method is robust against irregular shading and vein deformations due to posture changes. At first, the authors apply a technique originally proposed by Yang and Yang [50] for enhancing the vein images. Then a minimum-curvature map is calculated from the enhanced vein images based on Eigenvalue analysis. The feature point locations are determined from this curvature image (smaller Eigenvalue) at any point where the vein shape is non-linear. The feature descriptor takes the vein shape around the key-point location into account and is extracted from the so-called vein pattern map (larger Eigenvalue). The feature vector contains a quantification of the different vein directions inside a variable-sized window around the key-point location. The descriptor is normalised with the help of a finger shape model in a way that the descriptor area becomes smaller the closer the key-point location is to the finger boundaries. The authors employ a deformation tolerant matching strategy by using non-rigid registration. At first, the correspondences between the key-points in the two images for comparison are found. These correspondences are filtered using a local and global histogram technique based on the relative distances between the matched key-points. After this filtering step, the key-point coordinates of one of the involved feature vectors are transformed by applying a non-rigid transformation based on an outlier-robust thin-plate spline model as proposed in [37]. Afterwards, the correspondences between the adjusted key-points are determined again. These updated correspondences are filtered by a comparison of the descriptor distances with fixed thresholds. The final comparison score is determined as the ratio of the matched points and the sum of the number of detected key-points in both images. The authors claim that their proposed method is tolerant against several different types of finger posture changes, e.g. longitudinal finger rotation, translations and bending of the finger.
SIFT with Additional Key-Point Filtering (SIFT)
Key-point based techniques try to use information from the most discriminative points as well as considering the neighbourhood and context information of these points by extracting key-points and assigning a descriptor to each key-point. A SIFT [24]-based technique with additional key-point filtering as described in our previous work [14] is implemented in the vein recognition framework too. The key-point detector and descriptor is the default SIFT one, provided by the vl_featFootnote 12 MATLAB implementation. To suppress the unwanted finger outline information, the key-points located within a certain area around the finger’s outline are filtered (removed). The size of this area can be adjusted by a threshold.
SURF with Additional Key-Point Filtering (SURF)
Similar to SIFT, SURF is another general purpose key-point extractor and descriptor proposed by Bay et al. [5]. SURF is designed to be faster than SIFT while maintaining the robustness and description properties of SIFT. Our framework includes an OpenSURF [7] based vein feature extractor implementation with additional key-point filtering as for SIFT (described in [14]).
Finger Vein Recognition based on Deformation Information (Meng18)
The technique proposed by Meng et al. [29] is based on the observation that regular deformations, corresponding to posture changes, can only be present in genuine vein patterns. Thus, they incorporate pixel-based 2D displacements that correspond to these deformations during comparison. These displacements are determined with the help of a DenseSIFT descriptor-based approach. DenseSIFT extracts SIFT [24] descriptors at fixed points, defined by a dense grid. For each pixel, an 128-dimensional feature vector is extracted. Each pixel’s feature vector is compared against all feature vectors of the second image to find its nearest neighbour. Afterwards, a displacement matrix recording the pixel-wise displacements of all matched feature vectors is created. The displacement uniformity, calculated from displacement matrix, resembles the final comparison score.
Local Binary Patterns Based (LBP)
LBP has been successfully used in many different kinds of image feature extraction, also in finger vein recognition [23]. It is implemented as another representative of a binarisation-type feature extraction scheme. LBP compares the grey level of a centre pixel to its neighbouring pixels. Each pixel’s greyscale value is then represented by the corresponding binary code resulting from the comparison with its neighbourhood. This features can either be used directly during comparison or a histogram-based feature vector is created based on the LBP features. Therefore a histogram of the LBP values per input vein image is created and stored as a feature vector, which is then used during the comparison.
Phase Only Correlation (POC)
As the name suggests, POC [25] uses only the phase information in the frequency domain to compare two images. At first, the 2D Discrete Fourier Transform of both images which are to compare is calculated. Each of the coefficients can be represented by amplitude and phase. Then the cross-phase spectrum of the two images is determined. The POC function is defined as the 2D inverse Discrete Fourier Transform of the cross-phase spectrum. To suppress non-meaningful high-frequency components, a band-limited version of the cross-phase spectrum is calculated, by only including coefficients within a given window, which is centred at the correlation peak. The window size can be defined by two thresholds, a window width and window height, corresponding to the frequency band to be included. This variant is denoted as BLPOC (band-limited phase only correlation).
Morphological Post-Processing Options
Several morphological operations can be optionally applied after the feature extraction to enhance the feature images and remove noise. These operations include Skeletonisation, Thinning, Bridging, Fill, Shrink, Spur, Clean, Area Open, Inverse Area Open, Dilation, Erosion, Top Hat, Bottom Hat, Open and Close. Most of these operations require at least one additional parameter. Their implementation is based on the morphological image processing operations provided by MATLAB.
4.4 Comparison
This subsection lists the included comparison schemes, starting by two general purpose ones for binary templates and histogram data, followed by feature-type specific ones.
Miura Matcher (Miura)
For comparing the binary vein features we included the approach proposed by Miura et al. in [31, 32]. This approach is basically a simple correlation measure between an input and a reference image. Finger- as well as hand vein images are not registered to each other and only coarsely aligned (rotation is compensated for finger vein images) in general. Thus, the correlation between the input image and trimmed as well as in x- and y-direction shifted versions of the reference image is calculated. The maximum of these correlation values is normalised and then used as the final comparison score. The output score is a similarity score in the range of [0, 0.5] where 0.5 means a perfect match. This comparison scheme is selected by the comparison type Miura.
Histogram Comparison (Histogram)
Mainly for the comparison of the LBP histogram-based features, but not restricted to these features, we included several common histogram comparison metrics: histogram intersection distance, Chi-squared distance, Quadratic Form distance, Bhattacharyya distance, Jensen–Shannon divergence, Kolmogorov–Smirnov distance and Kullback–Leibler divergence. All those distance metrics can be selected if the comparison type is set to Histogram.
Feature-Type Specific Comparison
Several feature extraction schemes like DTFPM [27], ASAVE [51], POC [25], SIFT [14], Meng et al. [29] and others require a specific comparison scheme that can only be used with this particular kind of feature. Of course, all those feature specific comparison schemes are included as well and they are selected automatically if one of those feature types is set.
4.5 Comparison/Evaluation Protocols
In order to calculate the False Match Rate (FMR) and the False Non-Match Rate (FNMR) as defined by the ISO/IEC 19795-1 standard [39] prior to determining further performance numbers like the Equal Error Rate (EER), several different protocols are included. \(n_{gen}\) is the number of genuine comparisons, \(n_{imp}\) the number of impostor ones, \(n_{i}\)is the number of images/samples per hand/finger, \(n_{fh}\)is the number of hands/fingers that are captured per subject and \(n_{s}\)is the number of subjects.
FVC
This protocol is adopted from the fingerprint verification contests (e.g. the FVC2004 [26]). All possible genuine comparison are performed, i.e. each sample is compared against all remaining samples of the same biometric instance, i.e. finger/hand. No symmetric comparisons are performed, i.e. if the comparison [image A–image B] is performed, the comparison [image B–image A] is not performed as most comparison metrics are symmetric and both comparisons would result in the same score value. The number of genuine comparisons is
For the impostor comparisons, only the first sample of each finger/hand is compared against the first sample of all remaining fingers/hands. As with the genuine comparisons, no symmetric comparisons are performed. The number of impostor comparisons is
This protocol speeds up the comparison process as the number of impostor comparisons is greatly reduced while still ensuring that at least each finger/hand is compared against each other finger/hand once. The number of fingers/hands refers to the total number of fingers/hands in the dataset and not the number of fingers/hands per subject, e.g. if there are 60 subjects and 6 fingers in the dataset, then the number of fingers is 360. The above-mentioned number of genuine and impostor comparisons refers to the “probe only” comparison mode, i.e. that the gallery and probe directory are the same. In this case, one sample is not compared against itself. If the “gallery mode” comparison is used, i.e. the gallery directory contains different samples than the probe one, each probe sample is compared against the sample with the same subject/finger/sample ID within the gallery samples. Thus, the number of genuine and impostor comparisons is increased. As an example for the FVC protocol and 60 subjects, 6 fingers per subject and 4 images per finger (UTFVP dataset), the number of genuine comparisons is
and the number of impostor comparisons is
FVC (short)
An even shorter version of the FVC protocol in terms of the number of comparisons that are performed is included as well. In this version, the same number of genuine comparisons as for the FVC protocol is performed, but the number of impostor comparisons is further reduced. For the impostor comparisons, only the first sample of each finger/hand per subject is compared with the first sample of the finger/hand with the same ID for all other subjects, resulting in:
impostor comparisons.
Full
This protocol simply performs all possible comparisons without doing any symmetric ones, i.e.
genuine comparisons and
impostor comparisons are performed.
Ton
Ton and Veldhuis used a reduced version of the “Full” protocol in their paper about UTFVP finger vein dataset [45]. As they used about \(10\%\) of the dataset to tune the parameters of the recognition schemes, they excluded these samples from the subsequent performance evaluation. In order to do so, they skipped the first finger of the first subject, the second finger of the second subject, the first finger of the seventh subject and so on. This protocol is included in our vein recognition framework too, mainly to obtain recognition performance results that can be directly compared to the ones of the original UTFVP paper.
4.6 Performance Evaluation Tools
To evaluate the recognition performance of the different schemes on a particular dataset, several performance figures and plots can be generated by the vein recognition framework automatically. All these figures and plots are based on the genuine and impostor comparison scores that have been calculated according to one of the above-mentioned protocols.
EER/FMR100/FMR1000/ZeroFMR Determination
These basic numbers are commonly used to quantify the performance of biometric recognition schemes. The EER, is the point where the FMR and the FNMR are equal. The FMR100 (the lowest FNMR for FMR = \(1\%\)), the FMR1000 (the lowest FNMR for FMR = \(0.1\%\)) and the ZeroFMR (the lowest FNMR for FMR = \(0\%\)) are included as well. Besides these, the FNMR1000 (the lowest FMR for FNMR = \(0.1\%\)) and the ZeroFNMR (the lowest FMR for FNMR = \(0\%\)) are included too.
FMR Versus FNMR/ROC/DET Plots
Besides the performance numbers, the vein recognition framework does provide the most common used plots to indicate the recognition performance, which is the FMR versus FNMR plot, the ROC and the DET plot. According to the ISO/IEC 19795-1 standard [39], the ROC is a plot of the rate of false positives (i.e. impostor attempts accepted) on the x-axis against the corresponding rate of true positives (i.e. genuine attempts accepted) on the y-axis plotted parametrically as a function of the decision threshold. TheDET plot is a modified ROC curve which plots error rates on both axes (false positives on the x-axis and false negatives on the y-axis). Figure 4.4 shows a schematic ROC and DET plot.
4.7 Feature and Score-Level Fusion
The vein recognition framework contains tools and methods to perform feature and score-level fusion. A detailed description of the included feature-level fusion methods can be found in [10]. Regarding score-level fusion, several simple fusion schemes are included. Score normalisation is a vital point for score-level fusion. Consequently, several widely used score nomalisation schemes are included too. Below is a list of the included feature-level and score-level fusion schemes as well as the score normalisation schemes:
-
Feature-level fusion
-
Weighted mean and weighted sum
-
Majority vote
-
STAPLE, STAPLER, COLLATE, Spatial STAPLE provided by the MASI Fusion Toolkit [62].
-
-
Score-level fusion
-
Sum, Product, Minimum, Maximum, Mean, Median
-
Weighted versions of the above mentioned.
-
-
Score normalisation
-
Min-Max
-
Z-Score
-
Tanh
-
Median-MAD
-
Double-Sigmoid
-
Adaptive score normalisation for multimodal biometric systems [19].
-
These fusion and normalisation schemes can easily be extended by providing additional MATLAB implementations of the respective schemes and adding them to the source files of the vein recognition framework.
5 Experimental Example
This section gives an experimental baseline example, which should serve as a starting point for our vein recognition framework that is easy to use and reproduce. In the following, the used dataset, vein processing methods and the test protocol are described.
5.1 Dataset and Experimental Set-Up
This experimental example is conducted at the University of Twente Finger Vascular Pattern Database (UTFVP) [45]. This database consists of 1440 finger vein images, captured from 60 subjects, with 6 fingers (index, middle and ring finger of both hands) and 4 images per finger. The images are stored in the png format, have a resolution of \(672\times 380\) pixels and a density of 126 pixels/cm. The width of the visible blood vessels in the images is between 4 and 20 pixels.
The whole example can be run by using the runBookChapterExample.m script which is located inside the Tests directory. Only the dataset path in the script file has to be adjusted to point to the location of the UTFVP files.
The employed processing chain consists of the following components:
-
Preprocessing: at first the finger is segmented and the background region is masked out by using the LeeRegion [23] method with a filter width of 40 pixels and a filter height of 4 pixels. Afterwards, the finger is normalised using the HuangNormalise [8] approach. Then CLAHE [55] with a clip limit of 0.01 is applied, followed by the enhancement proposed by Zhang et al. [53] with a Gabor filter bandwidth of 1.12 and a Gabor filter Sigma of 3. Finally, the image is resized to half of its original size.
-
Feature extraction: two different feature extraction methods are employed. The first one is Miura’s Maximum Curvature method (MC) [32] with a Sigma of 2.5. The second one is Choi’s Principal Curvature method (PC) [6] with a Sigma of 2.5 and a Threshold of 1.5 plus an additional morphological post-processing using AreaOpen and InverseAreaOpen with 30 iterations each.
-
Comparison: The Miura Matcher [32] with a horizontal shift of 80 pixels, a vertical shift of 30 pixels and a rotational shift of 2\(^\circ \) in combination with the FVC evaluation protocol [26] is used during comparison of the extracted vein features. Based on the number of images contained in the UTFVP dataset, this results in 3600 genuine comparisons and 64620 impostor ones.
-
Performance evaluation: The recognition performance is evaluated using the EER, the FMR100, the FMR1000 and the ZeroFMR as well as the corresponding DET and ROC plots, which are shown in the following.
5.2 Experimental Results
Figure 4.5 shows an example image of the UTFVP dataset (first column) and the same image after vein region masking (second column) and after the applied preprocessing (third column). The fourth and rightmost column shows the extracted MC and PC features, correspondingly. The performance evaluation results are listed in Table 4.1 for both, the MC and PC features. MC performed overall better with an EER of \(0.37\%\) while PC achieved and EER of \(0.92\%\). The FMR values follow the same trend as the EER ones. The ROC plot is shown in Fig. 4.6 left and the DET plot in Fig. 4.6 right. Note that this example should serve as a starting point and is only for demonstrating the capabilities and basic workflow of the PLUS OpenVein Toolkit. Thus, the performance of the two feature extractors and the whole tool chain is not optimised to achieve the best possible recognition performance.
6 Conclusion and Future Work
In order to conduct research in vein recognition, two important things are necessary: A suitable and available dataset and an implementation of a vein processing tool chain, including preprocessing, feature extraction, comparison and performance evaluation. In this chapter, we presented a new open-source vein recognition framework, called PLUS OpenVein Toolkit, which implements a full vein recognition tool chain. This framework is implemented in MATLAB and contains various preprocessing, feature extraction and comparison methods as well as evaluation protocols and performance evaluation tools. Moreover, it includes feature- and score-level fusion functions. It supports many widely used finger- and hand vein datasets out of the box and it is easy to add support for new datasets. The source code of the framework is available free of charge for research and non-commercial purposes.Footnote 13 This chapter explained the basic structure of the framework, described the included vein processing and evaluation schemes and gave an experimental use case example. A more detailed description of all the individual parts is available with the source code. We also established a git repositoryFootnote 14 where every interested researcher is invited to contribute to our vein recognition framework by providing additional vein processing schemes and helping us to improve the code.
The framework is still being extended and improved. We plan to include additional preprocessing and feature extraction schemes as well as to optimise the currently implemented ones for runtime performance. Furthermore, we will include a special version of the FVC protocol, tailored to finger vein recognition, which distinguishes between different fingers (e.g. index vs. pinky one) during the comparisons. This enables an in-depth evaluation of possible differences between fingers of different size (the pinky finger is much smaller in diameter than the thumb). Finally, we plan to port the whole toolkit to either Python or C++ to get rid of the limitations imposed by MATLAB and to improve the runtime performance.
Notes
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
Can be downloaded at: http://www.wavelab.at/sources/OpenVein-Toolkit/.
- 9.
- 10.
The source code is hosted on our gitlab server: https://gitlab.cosy.sbg.ac.at/ckauba/openvein-toolkit.
- 11.
Supported means that the files are read and partitioned into subjects/fingers/samples correctly but not necessarily that optimised settings files for each single one of them exist.
- 12.
- 13.
Can be downloaded at: http://www.wavelab.at/sources/OpenVein-Toolkit/.
- 14.
References
Anjos A, Günther M, de Freitas Pereira T, Korshunov P, Mohammadi A, Marcel S (2017) Continuously reproducing toolchains in pattern recognition and machine learning experiments. In: International conference on machine learning (ICML). http://publications.idiap.ch/downloads/papers/2017/Anjos_ICML2017-2_2017.pdf
Anjos A, Shafey LE, Wallace R, Günther M, McCool C, Marcel S (2012) Bob: a free signal processing and machine learning toolbox for researchers. In: 20th ACM conference on multimedia systems (ACMMM), Nara, Japan (2012). https://publications.idiap.ch/downloads/papers/2012/Anjos_Bob_ACMMM12.pdf
Asaari MSM, Suandi SA, Rosdi BA (2014) Fusion of band limited phase only correlation and width centroid contour distance for finger based biometrics. Expert Syst Appl 41(7):3367–3382
Bandara AMRR, Rajarata KASHK, Giragama PWGRMPB (2017) Super-efficient spatially adaptive contrast enhancement algorithm for superficial vein imaging. In: 2017 IEEE international conference on industrial and information systems (ICIIS), pp 1–6. https://doi.org/10.1109/ICIINFS.2017.8300427
Bay H, Tuytelaars T, Van Gool L (2006) Surf: speeded up robust features. In: European conference on computer vision. Springer, pp 404–417
Choi JH, Song W, Kim T, Lee SR, Kim HC (2009) Finger vein extraction using gradient normalization and principal curvature. Proc SPIE 7251:9. https://doi.org/10.1117/12.810458
Evans C (2009) Notes on the opensurf library. Technical report CSTR-09-001, University of Bristol
Huang B, Dai Y, Li R, Tang D, Li W (2010) Finger-vein authentication based on wide line detector and pattern normalization. In: 2010 20th international conference on pattern recognition (ICPR). IEEE, pp 1269–1272
Kauba C (2015) Impact of sensor ageing on finger-image based biometric recognition systems. Master’s thesis (2015)
Kauba C, Piciucco E, Maiorana E, Campisi P, Uhl A (2016) Advanced variants of feature level fusion for finger vein recognition. In: Proceedings of the international conference of the biometrics special interest group (BIOSIG’16), Darmstadt, Germany, 12 pp
Kauba C, Prommegger B, Uhl A (2018) Focussing the beam—a new laser illumination based data set providing insights to finger-vein recognition. In: Proceedings of the IEEE 9th international conference on biometrics: theory, applications, and systems (BTAS2018). Los Angeles, California, USA, 9 pp
Kauba C, Prommegger B, Uhl A (2018) The two sides of the finger—an evaluation on the recognition performance of dorsal vs. palmar finger-veins. In: Proceedings of the international conference of the biometrics special interest group (BIOSIG’18), Darmstadt, Germany, 8 pp
Kauba C, Prommegger B, Uhl A (2019) PLUS OpenVein—an available fully open-source modular multi-purpose finger-vein scanner. In: Uhl A, Busch C, Marcel S, Veldhuis R (eds) Handbook of vascular biometrics. Springer Science+Business Media, Boston, MA, USA, pp 77–112
Kauba C, Reissig J, Uhl A (2014) Pre-processing cascades and fusion in finger vein recognition. In: Proceedings of the international conference of the biometrics special interest group (BIOSIG’14). Darmstadt, Germany, 12 pp
Kauba C, Uhl A (2015) Robustness evaluation of hand vein recognition systems
Kauba C, Uhl A (2015) Sensor ageing impact on finger-vein recognition
Kauba C, Uhl A (2018) Robustness of finger-vein recognition
Kauba C, Uhl A (2018) Shedding light on the veins—reflected light or transillumination in hand-vein recognition
Khalifa AB, Gazzah S, BenAmara NE (2013) Adaptive score normalization: a novel approach for multimodal biometric systems. World Acad Sci Eng Technol Int J Comput Sci Eng 7(3):882–890
Kirchgasser S, Kauba C, Uhl A (2019) Towards understanding acquisition conditions influencing finger-vein recognition
Kono M (2000) A new method for the identification of individuals by using of vein pattern matching of a finger. In: Proceedings of the fifth symposium on pattern measurement, Yamaguchi, Japan, 2000, pp 9–12
Kumar A, Zhou Y (2012) Human identification using finger images. IEEE Trans Image Process 21(4):2228–2244
Lee EC, Lee HC, Park KR (2009) Finger vein recognition using minutia-based alignment and local binary pattern-based feature extraction. Int J Imaging Syst Technol 19(3):179–186
Lowe DG (1999) Object recognition from local scale-invariant features. In: Proceedings of the seventh IEEE international conference on computer vision (CVPR’99), vol 2. IEEE, pp 1150–1157
Mahri N, Suandi SAS, Rosdi BA (2010) Finger vein recognition algorithm using phase only correlation. In: 2010 International workshop on emerging techniques and challenges for hand-based biometrics (ETCHB). IEEE, pp 1–6
Maio D, Maltoni D, Cappelli R, Wayman JL, Jain AK (2004) FVC2004: third fingerprint verification competition. In: ICBA, LNCS, vol 3072. Springer, pp 1–7
Matsuda Y, Miura N, Nagasaka A, Kiyomizu H, Miyatake T (2016) Finger-vein authentication based on deformation-tolerant feature-point matching. Mach Vis Appl 27(2):237–250
McCann J (1999) Lessons learned from mondrians applied to real images and color gamuts. In: Color and imaging conference, vol 1999. Society for Imaging Science and Technology, pp 1–8
Meng X, Xi X, Yang G, Yin Y (2018) Finger vein recognition based on deformation information. Sci China Inf Sci 61(5)
Mirmohamadsadeghi L, Drygajlo A (2014) Palm vein recognition with local texture patterns. Iet Biom 3(4):198–206
Miura N, Nagasaka A, Miyatake T (2004) Feature extraction of finger-vein patterns based on repeated line tracking and its application to personal identification. Mach Vis Appl 15(4):194–203
Miura N, Nagasaka A, Miyatake T (2007) Extraction of finger-vein patterns using maximum curvature points in image profiles. IEICE Trans Inf Syst 90(8):1185–1194
Prommegger B, Kauba C, Uhl A (2018) Longitudinal finger rotation—problems and effects in finger-vein recognition
Prommegger B, Kauba C, Uhl A (2018) Multi-perspective finger-vein biometrics
Prommegger B, Kauba C, Uhl A (2019) A different view on the finger—multi-perspective score level fusion in finger-vein recognition. In: Uhl A, Busch C, Marcel S, Veldhuis R (eds) Handbook of vascular biometrics. Springer Science+Business Media, Boston, MA, USA, pp 395–434
Raghavendra R, Venkatesh S, Raja K, Busch C (2018) A low-cost multi-fingervein verification system. In: Proceedings of international conference on imaging systems and techniques (IST 2018), Karkow, Poland
Rohr K, Fornefett M, Stiehl HS (1999) Approximating thin-plate splines for elastic registration: integration of landmark errors and orientation attributes. In: biennial international conference on information processing in medical imaging. Springer, pp 252–265
Shi Y, Yang J, Yang J (2012) A new algorithm for finger-vein image enhancement and segmentation. Inf Sci Ind Appl 4(22):139–144
For Standardization, I.O. (2006) Iso/iec is 19795-1. Information technology—biometric performance testing and reporting—Part 1: Principles and framework
Starck JL, Fadili J, Murtagh F (2007) The undecimated wavelet decomposition and its reconstruction. IEEE Trans Image Process 16(2):297–309
Syarif MA, Ong TS, Teoh AB, Tee C (2017) Enhanced maximum curvature descriptors for finger vein verification. Multimedia Tools Appl 76(5):6859–6887
Tome P, Marcel S (2015) On the vulnerability of palm vein recognition to spoofing attacks. In: The 8th IAPR international conference on biometrics (ICB). http://publications.idiap.ch/index.php/publications/show/3096
Tome P, Vanoni M, Marcel S (2014) On the vulnerability of finger vein recognition to spoofing. In: IEEE international conference of the biometrics special interest group (BIOSIG). http://publications.idiap.ch/index.php/publications/show/2910
Ton B (2012) Vascular pattern of the finger: biometric of the future? sensor design, data collection and performance verification. Master’s thesis, University of Twente
Ton B, Veldhuis R (2013) A high quality finger vascular pattern dataset collected using a custom designed capturing device. In: International conference on biometrics, ICB 2013. IEEE. http://doc.utwente.nl/87790/
University of Reading: PROTECT Multimodal DB Dataset (2017). http://projectprotect.eu/dataset/
Xie SJ, Lu Y, Yoon S, Yang J, Park DS (2015) Intensity variation normalization for finger vein recognition using guided filter based singe scale retinex. Sensors 15(7):17089–17105
Yang J, Shi Y (2012) Finger-vein roi localization and vein ridge enhancement. Pattern Recogn Lett 33(12):1569–1579
Yang J, Shi Y, Yang J (2012) Finger-vein image restoration based on a biological optical model. In: New trends and developments in biometrics. InTech
Yang J, Yang J (2009) Multi-channel gabor filter design for finger-vein image enhancement. In: Fifth international conference on image and graphics, 2009. ICIG’09. IEEE, pp 87–91
Yang L, Yang G, Yin Y, Xi X (2017) Finger vein recognition with anatomy structure analysis. IEEE Trans Circuits Syst Video Technol
Yin Y, Liu L, Sun X (2011) Sdumla-hmt: a multimodal biometric database. Biomet Recogn 260–268
Zhang J, Yang J (2009) Finger-vein image enhancement based on combination of gray-level grouping and circular gabor filter. In: ICIECS 2009 international conference on information engineering and computer science, 2009. IEEE, pp 1–4
Zhao J, Tian H, Xu W, Li X (2009) A new approach to hand vein image enhancement. In: Second international conference on intelligent computation technology and automation, 2009. ICICTA’09, vol 1. IEEE, pp 499–501
Zuiderveld K (1994) Contrast limited adaptive histogram equalization. In: Heckbert PS (ed) Graphics gems IV. Morgan Kaufmann, pp 474–485
Online Resources
Bandara R, Kulathilake H, Giragama MP (2019) Speeded up adaptive contrast enhancement (SUACE). https://github.com/ravimalb/suace/. Accessed 23 Jan 2019
Bankhead P, Scholfield CN, McGeown JG, Curtis TM (2019) ARIA—Automated Retinal Image Analyzer. http://petebankhead.github.io/ARIA/. Accessed 23 Jan 2019
Cermelj P (2019) INIFILE—INI file reading and writing. https://de.mathworks.com/matlabcentral/fileexchange/2976-inifile. Accessed 23 Jan 2019
Funt B, Ciurea F, McCann J (2019) Retinex in Matlab. http://www.cs.sfu.ca/~colour/publications/IST-2000/. Accessed 23 Jan 2019
van der Heijden F (2019) Image edge enhancing coherence filter toolbox. https://de.mathworks.com/matlabcentral/fileexchange/25449-image-edge-enhancing-coherence-filter-toolbox. Accessed 23 Jan 2019
Mayoue A (2019) BioSecure tool—performance evaluation of a biometric verification system. http://svnext.it-sudparis.eu/svnview2-eph/ref_syst/Tools/PerformanceEvaluation/. Accessed 23 Jan 2019
Medical Image Analysis and Statistical Interpretation (MASI) group at Vanderbilt University (2019) MASI label fusion. https://www.nitrc.org/projects/masi-fusion/. Accessed 23 Jan 2019
Schauerte B (2019) Histogram distances https://de.mathworks.com/matlabcentral/fileexchange/39275-histogram-distances. Accessed 23 Jan 2019
Ton B (2019) Finger region localisation. https://de.mathworks.com/matlabcentral/fileexchange/35752-finger-region-localisation. Accessed 23 Jan 2019
Ton B (2019) Miura et al vein extraction methods. https://de.mathworks.com/matlabcentral/fileexchange/35716-miura-et-al-vein-extraction-methods. Accessed 23 Jan 2019
Ton B (2019) Simple finger normalisation. https://de.mathworks.com/matlabcentral/fileexchange/35826-simple-finger-normalisation. Accessed 23 Jan 2019
Ton B (2019) Wide line detector. https://de.mathworks.com/matlabcentral/fileexchange/35754-wide-line-detector. Accessed 23 Jan 2019
Vedaldi A, Fulkerson B (2019) VLFeat—VisionLab features library. http://www.vlfeat.org/. Accessed 23 Jan 2019
Xiong G (2019) Local adaptive thresholding. http://de.mathworks.com/matlabcentral/fileexchange/8647-local-adaptive-thresholding. Accessed 23 Jan 2019
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 700259, project PROTECT—Pervasive and UseR Focused BiomeTrics BordEr ProjeCT.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2020 The Author(s)
About this chapter
Cite this chapter
Kauba, C., Uhl, A. (2020). An Available Open-Source Vein Recognition Framework. In: Uhl, A., Busch, C., Marcel, S., Veldhuis, R. (eds) Handbook of Vascular Biometrics. Advances in Computer Vision and Pattern Recognition. Springer, Cham. https://doi.org/10.1007/978-3-030-27731-4_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-27731-4_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27730-7
Online ISBN: 978-3-030-27731-4
eBook Packages: Computer ScienceComputer Science (R0)