Abstract
Nanobiosensors (NBSs) are a class of chemical sensors which are sensitive to a physical or chemical stimulus (heat, acidity, metabolism transformations) that conveys information about vital processes. NBSs detect physiological signals and convert them into standardized signals, often electrical, to be quantified from analog to digital. NBSs are classified according to the transducer element (electrochemical, piezoelectric, optical, and thermal) in accordance with biorecognition principle (enzyme recognition, affinity immunoassay, whole sensors, DNA). NBSs have varied forms, depending on the degree of interpretation of natural processes in plants. Plant nanobionics uses mathematical models based on qualitative and less quantitative records. NBSs can give information about endogenous concentrations or endogenous fluxes of signaling molecules (phytohormones). The properties of NBSs are temporal and spatial resolution, the ability of being used without significantly interfering with the system. NBSs with the best properties are the optically genetically coded NBSs, but each NBS needs specific development efforts. NBS technologies using antibodies as a recognition domain are generic and tend to be more invasive, and there are examples of their use in plant nanobionics. Through opportunities that develop along with technologies, we hope that more and more NBSs will become available for plant nanobionics. The main advantages of NBSs are short analysis time, low-cost tests and portability, real-time measurements, and remote control.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
12.1 Introduction
Plants are a fascinating research topic if we relate to environmental stress. Because they are physically stuck in specific spots, the plants have to handle in that site, regardless of the environmental conditions. Moving to another place is not an option. But what plants can do is to modify the internal “environment,” and plants are “true masters” of manipulating their metabolism to deal with environmental disturbances. This feature is one of the reasons why plants are useful in various research; we can rely on them as “sensitive indicators” of environmental changes, even in completely new environments. In the absence of normal conditions, plants cannot use the classical pathways of metabolism, so they need to identify other solutions. This is what happens when plants adjust their metabolism for regulating gene activation, thus producing more or less proteins that are useful or not in the new environmental conditions. The different parts of the plant come with their own genetic regulation strategies.
A number of genes that are involved in the creation and remodeling of cell walls are activated differently in growing plants. Other genes with a role in identifying light, which are normally active on the leaves, are active at the root level. In leaves, many genes associated with transmission of hormonal information are suppressed, and genes associated with insect protection are more active. These trends are also seen in the (higher) number of proteins involved in message transmission, cell wall metabolism, and plant protection. These patterns of gene and protein functioning indicate that in unfavorable conditions, the plants respond by weakening the cell walls and creating new ways to understand the environment. It is possible to monitor changes in the genes in real time by labelling certain proteins with fluorescent elements. Plants modified with fluorescent proteins can give useful information about how they respond to the environment. These modified plants act as biological sensors (NBSs).
Specialized cameras and microscopes allow us to monitor how the plant uses these fluorescent proteins (Hamers et al. 2014). Chemical or biological NBS functions on the principle of signal emission (voltage or electrical, photonic) in response to a chemical reaction involve a chemical or biological receptor, R (macrocyclic ligand, antibody enzyme), that binds to a specific target molecule of a sample to be studied, the analyte, A.
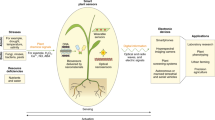
Signal transmission is achieved by coupling with a transducer T that interfaces NBS processes with the processing–transform unit in a measurable signal. Analysis of signals in plant nanobionics aims at processing signals recorded by measurements in order to extract the maximum of useful information for diagnostics and monitoring. An NBS is a measurement tool for physiological measures which transforms them into measurable signals. As shown in Table 12.1, there are many areas with important applications for chemical and biological sensors, from continuous monitoring of chemical processes to plant nanobionics (He et al. 2016).
Nanotechnology is the ability to build objects by assembling atoms at a time in well-defined sequences. With the tools provided by nanotechnology, genetic engineering, and proteomics, “bottom-up” objects can be built, meaning assembling atoms and molecules into structures. To do this, other types of nanometer-sized instruments and devices are needed. To make macroscopic objects, billions of these small devices (assemblies) are needed. To reproduce these types of tools, we need a self-replication method or replication tools (replicators).
Essential components in a sensory structure have unique meanings due to miniaturization and large-scale integration into integrated circuits, making themselves chips with a strong interfacing of the analyte–receiver reaction processes. By consensus it has been established that the term should be reserved for use as a sensor that incorporates biological elements such as enzymes, antibodies, nucleic acids, microorganisms, or cells. Table 12.1 shows the interferences of the NBS chemosensor applications during their evolution.
NBSs will be defined as a compact analytical device incorporating a biologic sensing element or biologic derivative integrated with a physicochemical transducer. The goal of NBSs is to produce an analog or digital electronic signal that is proportional to the concentration of a single analyte or a group of analytes.
These devices are mostly used in genetic engineering in agriculture, where it is necessary to know the mechanisms of reaction and the affinity of enzymes and microorganisms for different substrates of interest and signaling molecules. A biochip is a device that contains a structure of individual sensory elements (NBSs) interconnected by functions and recognition specifications, integrated on a chip. The number of NBSs on a chip may be of the order of 106 units. In this degree of integration, a set of distinct tests can be performed extremely quickly and efficiently. In contrast to microchips, biochips are not electronic structures (they contain different electronic structures coupled to nanobiosensory elements). Each NBS can be thought of as a “microreactor” that performs a specific chemical reaction with an analyte. NBSs from biochips can be designed to detect a wide variety of analytes, including DNA, antibodies, proteins, and biomolecules. The advantages are multiple: sensors can be produced in batches or sequences that can be assembled in parallel or serial, providing a high manufacturing yield; sensors can be assembled on very small areas with reduced distances between them; 3D structures can be generated providing high signals besides 2D structures; any type of biochemical reaction may be incorporated; NBSs can be produced separately and subsequently assembled according to the specificity and nature of the application.
The important features regardless of industry or technology are selectivity, sensitivity, and stability in the design of sensory systems integrated with structures and arrangements of sensory elements (Wan Salim et al. 2013). One of the integrated systems including rotational aseptic sampling is a robotic fluid and reusable electrodes formed by ink-jet printing injection system.
The system contains an enzyme electrode with immobilized GOx in gel, and the detection of hydrogen peroxide is carried out on a rhodinised carbon electrode (Rh coating). Although the enzyme electrode has stability and efficiency characteristics, the problem of automated sample monitoring and sampling in an integrated system requires multiparameter optimization due to reciprocal interferences. There is a requirement on specific domains (environment and genetic engineering) of highly performing integrated systems that work in in vivo conditions, such as dialysis, the use of biointerfaces, evanescent techniques, and atomic force microscopy to grasp in the depth of the biological phenomena (identifying and understanding the interaction of proteins). The in vivo exploitation of detection systems for both glucose and lactate was confirmed by the efficacy of using phospholipid copolymers and improving hemocompatibility.
Immunosensors provide an example for the development of integrated systems where microseparations, chromatographic methods, and electrochemical couplings with optical detectors are incorporated, which ultimately lead to a miniaturized system. There are examples where the level of integration and miniaturization becomes more pronounced (DNA–nanotube or biochips–biointerfaces). NBSs are expected to be widely used in plant nanobionics where physiological/biochemical parameters should be identified.
Advanced ink-jet technology has developed methods for analyzing nanoliter fractions on a three-dimensional NBS surface at a speed of 6 m/s. It is expected to produce one million NBSs/cm2 areas using photolithography, contact fingerprinting or self-assembling techniques, and adsorption/desorption under the laser beam that allows “writing” proteins on the surface to be analyzed with great precision. Laser techniques, MAPLE (Matrix Assisted Pulsed Laser Evaporation) or DW (Direct Writing) approached for immobilizing biological materials on substrates, are in the laboratory stage but have prospects for use as molecular imprinting methods (Potocký et al. 2014).
12.1.1 Sensitivity
Whether NBSs are individuals, in integrated systems, or areas of NBSs, all are characterized by unique parameters such as sensitivity and detection limit for a range of analytes. Trace detection of various analytes (indicators, additives, contaminants) with sufficient sensitivity and safety is the basic criteria of an NBS to be used. The detection limit in the laboratory is pushed to an atom when the atomic force microscopy is used. Thus, the enzymatic electrodes, studied and continuously perfected, use palliative such as concentrating the analyte of interest, which leads to major design and miniaturization difficulties.
NBSs for phenol vapors were identified, where the phenoloxidase was immobilized on a glycerol gel with a range of interdigitated electrodes. Phenol vapors are directly partitioned into the gel and oxidized to quinone. Signal amplification was improved by redox amplification of the quinone/catechol couple to obtain a reasonable sensitivity resulting in a detection limit of 30 ppb phenol. This principle can be extended to other carbon compounds up to the ppt (parts per trillion) limits. DNA structures have been studied as potential receptors.
Sandwich structures of liquid crystal dispersions and DNA-polycation complexes have been studied with relatively good success in identifying different analytes. The polycation with the role to maintain structural integrity of DNA and DNA-protamine complexes allows detection of hydrolysis of the trypsin enzyme to the detection limit of 10−14 M.
Elimination of the polycation leads to an increase in the distance between the two DNA strands resulting in the appearance of an intensive band in the circular dichroism spectrum due to the texture modification (Espinoza et al. 2014).
12.1.2 Stability
From a practical standpoint, the disadvantage is their inherent instability. Different strategies have been approached to improve longevity and preserve the structure of biological receptors. Immobilization in matrices by sol–gel technique for glucose NBSs is one of the strategies; fluorescence indicators are used: hexahydrate chloride (2,2′-bipyridyl) ruthenium (II) and 1-hydroxypyrene-3,6,8 trisulfonic acid.
In addition to the optical property improvement of the gel, the stability of the GOX enzyme has also improved. Other examples are the case of monooxygenase used in hydrocarbon detection; detection of organic halides with metalloporphyrins; and detection of carbon tetrachloride, haloalkane (perchloroethylene), and insecticides (DDT) (Kazakova et al. 2013).
12.1.3 Selectivity
Improving the selectivity of an NBS can be approached on two levels: through direct transducer–biological receiver interfacing to reduce interference and new receptors with improved affinity or new affinity capabilities. Selectivity is a key parameter that requires the performance of an NBS. Pyrroloquinoline is used as a mediator in a glucose oxidase enzyme electrode for the measurement of glucose in the raw or elaborated cavity.
Alternatively, the electrocatalytic detection of the reaction products resulting from the enzymatic reactions can be improved by chemically modified electrodes such as rhodinised electrodes or hexacyanoferrate modified carbon electrodes. Prussian blue is used to modify the electrode surface at amperometric detection of oxygenated water at both oxidation and reduction potentials for the enzyme electrode in lactate and glucose detection.
One solution is to identify redox centers of the enzyme via a molecular wire to perform the electron transfer to the electrode (enzymes linked by molecular wires), but the concerns have focused on immobilized mediators on different polymer chains. Molecular wires are regarded as intermediates in long-range electron transfer, consisting of two pyridine groups linked by thiophenes with different lengths. Such wires can be used in conjunction with self-assembly techniques to produce an isolated electrode that transfers electrons to predetermined molecular pathways (Jones et al. 2013).
12.2 The Biological Components of Nanobiosensors (NBSs)
An ideal NBS is a device that will detect an “analyte,” the subject under analysis and which is present in a given sample. Most samples also contain other analytes which may interfere with the NBS response. It must have a specific selectivity to identify the target analyte. It is necessary to design NBSs with selectivity for an analyte with the ability to discriminate the interferences produced by the other components in the analyzed sample.
Specific identification and selectivity capacity are the key components of molecular recognition. Molecular recognition is accomplished by the sensor component of a host molecule (host–chemoreceptor) that binds selectively to the “guest” target molecule/guest molecule that needs to be identified. For each “host–guest” system, there is a specific chemical reaction from the multitude of possible reaction channels. When the host–guest response was identified, the host molecule is immobilized/incorporated into NBSs, typically on a transducer-contacting membrane/contact electrode. Finally, a way to signal that the bindings/recognition event has occurred (transducer to transducer) has to be found (Rodríguez–Sevilla et al. 2014).
12.2.1 Principles of Molecular Recognition
One of the requirements for molecular recognition is the existence of groups or centers with specific reactivity in the host molecule that can “close” or bind ions, atoms, molecules, and biomolecules. All living organisms use enzymes, which are proteins that contain “pockets,” active centers, designed to recognize a specific analyte. This means that only a specific analyte is able to enter the enzyme pocket. Enzymes can be used in NBSs as host receptors with molecular recognition capability but are unstable. To design host molecules that can be used in an NBS, the following criteria are considered: the host molecule must be stable under the conditions in which it is to be used, must be able to selectively bind the analyte in the sample, must be able to be immobilized in a film/membrane that is in contact with the sample, must signal that a host–host binding response has occurred, and must ideally release the analyte after detection so the host is free to be reusable. Biological receptors include antibodies, membrane receptors, signaling molecules, enzymes, ribosomes, lectins, phytohormones, etc.
They bind analytes using “lock-and-key” molecular recognition mechanisms (key–lock, identification–immobilization). Biological receptors are not practical solutions for many applications because the specificity, sensitivity, and stability cannot be optimized. Artificial receivers are immobilization environments that can be optimized by molecular design for any type of application. Synthetic receptor design and synthesis are based on tools developed by proteomics and genetic engineering, producing recognition components that can respond to the occurrence and identification of metabolic deficiencies in plant nanobionics.
There are platforms and areas of artificial receptors based on combinatorial mathematical techniques, interface biology, and surface chemistry. They have induced the development of various artificial receptor environments with rapid and diversified selection for any target analyte. The current technique for producing synthetic receptors is called CARA (combinatorial array of receptor analysis) (Fang et al. 2015).
Supramolecular chemistry has developed a wide range of synthetic macrocycles. The most common feature for macrocycle classes is that they contain cavities that act as host pockets for guest molecules. The selectivity of the hosts can be done in “read mode” by varying the size of the preformed cavities. 12-crown-4 has a small cavity ideal for the binding of small ions such as Li+, while 18-crown-6 has a large cavity that fits better with larger ions such as K+.
The size of the cavities is important for the selectivity of the host, but the question remains what attracts an ion or molecule into a preformed cavity and which factors stabilize the host–guest complex. In enzymes, weak noncovalent interactions (hydrogen bonding, electrostatics, dipole–dipole, van der Waals, etc.) are used to link the guest into the enzyme pocket (interactions stabilize host–guest interaction). Macrocycles contain polar functionalities, capable of interacting with guests via hydrogen bonding, electrostatic interactions, and dipole–dipole interactions. It is desirable that bonding in the cavity is not strong, because it is important that the analyte, the guest, is released from the host after it has been detected and measured. Crown ethers and calixarene are ideal for bonding metal cations, based on the size of their cavity, but also on the high density of electrons present on the oxygen atoms in the cavity. The base compound selectively binds Li+ to other metallic cations; the modified version of the base macrocycle has a good selectivity for Na+. By synthetic modification it is possible to increase the capacity of the host cavity, and new functionalities can be introduced that will favor the binding of specific molecules and ions (da Silva et al. 2013).
Other modified calixarenes, which demonstrate this principle, are the group of tetraphosphine oxide of the calix[4]arene. By changing binding groups on the same template, calix[4]arene from esters in phosphorus hydrogen oxides selectivity changes from Na+ to Ca2+.
By increasing the number of repeatable units in esters and phosphorus hydrogen oxides at six, the cavity increases, and the selectivity changes in favor of the higher Cs+ and Pb2+ cations, respectively. Some host compounds have been developed for the selective detection of low molecular weight compounds. An example involves the use of the tetra (S-propanol) calix[4]arena containing four lateral chiral halves for selective differentiation of the phenylalanyl enantiomers. Other techniques of supramolecular chemistry may be involved in the synthesis of synthetic receptors that simulate the properties of enzymes. The basic structures that can be modified are porphyrins, semiconductor polymers of the tetrathiafulvalene (TTF) class, and PPVs. Other patterns can be considered modified polysaccharides. A linear archetype is the polyanilines containing the two types of redox states (Meyer et al. 2007).
12.2.2 Molecular Basics of Ag–Ab Interaction
Among the multiple NBS classifications, bioaffinity has a range of applications, and antigen–antibody interactions (Ag–Ab) play a role and are considered to be an instrument in the development of molecular recognition principles. In vivo Ag–Ab interactions are reversible. Factors that condition the Ag–Ab interaction are the structural complementarity between the antigenic determinant and the antibody combining site; this is the exclusive factor of the specificity of the reaction; the structural complementarity involves the conformational adaptation of the two reacting groups and was conceived in structural terms on the key–lock principle; the chemical complementarity of the reaction groups is the consequence of structural complementarity and signifies the entry into action of intermolecular forces that stabilize and consolidate the interaction of the two groups.
The formation of intermolecular bonds requires the existence of atomic groups sufficiently close to the two molecules. The distance between them is inversely proportional to the degree of complementarity. Although structural complementarity is not strictly obligatory, higher spatial matching is more conducive to interaction. It is expressed by the congruent of contact surfaces that provide intermolecular attraction forces that stabilize the complex.
The Ag–Ab interaction involves the following types of noncovalent bonds: H bonds, electrostatic forces, van der Waals linkages, and hydrophobic bonds. All are nonspecific forces of low value and their nature makes the reaction reversible. H bonds form when two atoms share an atomic H nucleus (one proton). The common proton is found between two atoms of N or O or between N and O. The H nucleus is covalently bonded to one of the two atoms (N or O). The H bond has a binding energy of 3–7 kcal/mol.
Intermolecular forces are involved in Ag–Ab complex formation. The action of these forces requires close contact between the two reaction groups. The H bonds result from the formation of an H bridge between two nearby atoms.
The electrostatic forces are due to the attraction of the ionic groups with opposite charges located at the periphery of the two protein chains. The van der Waals forces result from the interaction between different electron clouds, represented in the form of oscillating dipoles.
The van der Waals relationships, the weakest interaction forces, are active at very small distances between the reaction groups. The binding energy is 1–2 kcal/mol. Van der Waals’ links are not based on a permanent separation of electrical charges but on their fluctuations induced by the proximity of molecules. At intermolecular distance, instantaneous electric fields are formed, with a polarizing effect on neighboring molecules.
Between the nearby atoms, there is a mutual attraction force induced by fluctuating dipole load, which a dipole induces in the neighboring dipole (dispersion forces). Their intensity depends on the distance between the groups involved and is inversely proportional to the seventh power of the distance. Their value is optimal at 1–2 Å.
Hydrophobic bonds, which can contribute with half of the Ag–Ab binding force, are produced by the association of nonpolar and hydrophobic groups, whereby water molecules are excluded. The optimum distance between the reactive groups varies with the type of bond.
The electrostatic forces (coulomb or ionic) are the result of the attraction between atoms or groups of atoms with the opposite electric charge located on the two reacting groups: between a cation (Na+) and an anion (Cl−) or between COO− and NH3+ (Agrawal et al. 2012).
The binding energy of these forces is significant at very small (less than 100 Å) distances from the reaction groups. Exact juxtaposition of ions favors the action of these forces. The binding energy is 5 kcal/mol and varies inversely with the square of the distance between the two reaction groups (1/d2). Hydrophobic (or a polar) linkages occur between nonpolar (nonionized) groups in aqueous solutions and are the consequence of the tendency to exclude the ordered molecule of water molecules between the antigen and antibody molecules.
These linkages are favored by amino acids with a polar group that tends to associate, reducing the number of water molecules in their vicinity. By removing the water molecules between the reaction groups, the distance between the active sites decreases much, and the value of the stabilizing forces increases. Taken each one on their own, space complementarity or intermolecular forces are not sufficient to form stable relationships.
For the stability of the Ag–Ab interaction, both conditions are required. The higher the binding energy of the reactants, the stable the Ag–Ab complexes. The interaction of the antigen- and antibody-reactive groups is defined by two parameters: the affinity and avidity of the antibodies. Measurement of antibody affinity can be achieved by dialysis at equilibrium. The Ag–Ab interaction is reversible. Within the dialysis bag, the hapten is partially free and partially bound to the antibodies, depending on the affinity of the antibodies. Through the membrane of the dialysis bag, only the free hapten can be diffused, and its external concentration will equal the concentration of the free hapten inside the bag (Kersten and Feilner 2007).
Measurement of the concentration of the hapten in the dialysis bag allows for the calculation of the amount of antibody-bound hapten. The constant renewal of the buffer results in total dissociation and loss of hapten in the dialysis bag, which indicates the reversible nature of Ag–Ab binding. Affinity of antibodies measures the binding force between an antigenic determinant and the complementary binding site of a specific antibody. Affinity is the result of attraction and rejection forces that mediate the interaction of the two reactants.
The strength of these interactions is measured in the reaction between a monovalent antigen (hapten) and specific antibodies. A high affinity interaction requires perfect complementary structures, while the imperfect complementarity of the reaction groups causes a low affinity, since the attraction forces are active only at very small distances and are diminished by the rejection forces.
Complexes formed by antibodies with high affinity are rapidly eliminated from the circulation without adverse effects on renal function. The Ag–Ab interaction is permanently characterized by the formation and cancellation of various types of intermolecular bonds. In vivo, probably all Ag–Ab reactions are reversible, but secondary in vitro reactions (agglutination, precipitation), under the conditions of reagent balance, are irreversible (Sakamoto et al. 2018).
It is essential that the host–guest binding event (the receptor–analyte interaction, R–A) is detected. It is thus necessary to have a way of identifying and transducing the signal from the receptor–analyte interaction to the outside to be processed. It is generally defined as a transducer. The transducer must be in contact with the receiver or the diaphragm that immobilized the handset. Electrode and interfacial interactions are determinant in capturing the signal from an R–A interaction and transforming it into an electrical or photonic signal. There are ways of identifying the R–A event, collecting the signal and transducing it as an external signal. The way of identifying the signals and their transduction defines the type of NBSs.
This means immobilizing the receiver on an electrochemical transducer that measures a current (amperometric method) or a voltage (potentiometric ) between two electrodes. If R is immobilized on an optical component, then we will define optical NBSs (optical fiber, fluorescence, absorption, surface plasmon resonance (SPR)). For detection, at most electrochemical NBSs, it is necessary for the membranes containing the host molecule to be placed on a surface of an electrode that leads to an electrochemical response to the binding of the guest.
The approach works well when target analysts are loaded species such as metal cations. Neutral molecules cannot be detected from the point of view of electrochemical transduction, so the optical detection methods have been successfully used. A chiral host in calixarene contains naphthyl, fluorescent units. Upon binding to the guest, fluorescence is attenuated as a result of interaction between the naphthyl–phenyl groups in the host or analyte. The fluorescence attenuation is proportional to the concentration of the analyte. Optical methods are used because they offer greater sensitivity than electrochemical techniques. In the absence of the guest analyte, this compound does not exhibit fluorescence because the pyrenyl substituent cannot come in contact with the adjacent nitrophenyl substituent (and the fluorescence attenuation occurs due to their interaction).
However, in the presence of Na+ ions, fluorescence is observed, because the Na+ ion enters the cavity and binds to the oxygen in the phenoxy and carbonyl groups in the host. This binding induces a more rigid conformation by removing the nitrophenyl groups of pyrene to prevent fluorescence attenuation (Gaggeri et al. 2012).
12.2.3 Types of Biological Components
NBS is a bioelectronic analysis system that combines a transducer with a biological component that is in a specific interdependence. NBSs use biological systems with different levels of recognition of the substances to be determined. The first step in this interaction is the formation of the specific complex of the biologically active, immobilized substance R (the receptor, the substrate with the sensitive biological component) with the analyte A (often defined as the chemical signal). Table 12.2 summarizes specific patterns of NBSs in relation to the nature of the receptor and the chemical/biochemical signal.
There are two general classes of NBSs that are based on the bioaffinity response between R and A that alters the distribution of electrical charges that can be measured with specific transducers or consuming the substrate by a specific reaction. The biological constituent of the molecular recognition element (R) is represented by various active species that can be enzymes or enzymatic systems, antibodies (Ab) or antigens (Ag), receptors, populations of bacteria or eukaryotic cells, tissue fragments, and sometimes even signaling molecules. Analytes or substances that can be analyzed (A) are glucose or other sugars, amino acids, alcohols, lipids, and nucleotides.
They can be identified by their specific interaction, or their concentration can be measured by various methods. Both R and A represent distinct molecular species with high macromolecular specialization (antibodies, antigens, enzymes, receptors, etc.) or are complex systems (cells, tissues, etc.) (Kersten and Feilner 2007). After the active biological component, they can be grouped as follows:
-
Enzymatic NBSs: Enzymes are energy proteins characterized by their catalytic function. Modified substrate molecules lead to oxidation, reduction, and hydrolysis reactions that can be measured by enzymatic NBSs. Enzymatic NBSs produce a linear response depending on the substrate concentration.
-
Immunosensors: Antibodies are glycoproteins produced by the immune system when an external substance, antigen, is involved. It is theoretically possible to produce antibodies without identifying an antigen. An immunosensor is a high sensitivity NBS. The principle of operation is based on the Ag–Ab interaction of molecular recognition.
-
NBSs with receptors: The regularity of biological processes is ensured by high sensitivity molecular processes based on the specialization of structural proteins (receptors) capable of recognizing a number of physiological signals. This is the case for neurotransmitters, whose action is mediated through the presence of receptors in the plasma membrane, in sites or cell targets. Activation of the biologically active site is via the ion channels. The acetylcholine receptor is the first known receptor in neurotransmitting phenomena.
-
NBSs based on cells or tissues: Measurement of molecular species is not limited to interaction with the compounds to be analyzed; the transformations that occur can be measured as resulting products. It is desirable to operate with cell populations whose major metabolic pathways are known. Relevant is NBSs’ L-arginine, which associates populations of bacterial cells of Streptococcus faecium in combination with an ammonia electrode. Arginine is metabolized by microorganisms. It is difficult to obtain complex reactions outside the cellular structures. Similar to the use of cell populations as sensitive elements, fragments or parts of plant tissues may be used. The advantage is greater because there is no extra effort to keep the cells viable in a natural arrangement. For adenosine NBSs, a tissue biosensitive element has been proposed. For dopamine NBSs, specialists have focused on the pulp of banana fruit, considering that it has remarkable biocatalytic properties.
-
NBSs with redox proteins: The redox proteins are involved in biochemical processes such as cellular respiration and photosynthesis reaction (Kersten and Feilner 2007).
NBS catalysts use enzymes, microorganisms, or cells to catalyze a reaction with a target substance. NBSs own an affinity on using antibodies, receptors, and nucleic acids that bind to a target substance.
Reactions are quantified by electrochemical, optical, evanescence transducers, etc.
The main types of known redox proteins are cytochromes, containing iron in the prosthetic group, and cytochrome “c” is involved in the transfer of electrons into mitochondria; ferredoxins contain iron and sulfur ions in dimeric combinations of chloroplasts (2Fe–2S) ferredoxin and tetrameric combinations of bacterial ferredoxin 2 (2Fe–2S) involved in photosynthesis and transfer of fixed nitrogen ions, respectively; blue proteins contain copper linked to the smallest cysteine residue involved in a tetrahedral structure such as plastocyanin and azurin that mediates electron transfer in photosynthesis and possibly in nitrite reduction; flavoproteins, containing a prosthetic group and an organic conjugate, are involved in the transfer of proteins such as flavotoxins (Agrawal et al. 2012). These proteins play a role in nature, due to the location on their surface of the redox centers. The subtle architecture of molecules offers selectivity and specificity to these molecules in their interaction with other proteins or enzymes, such as the cytochrome c structure. Porphyrin iron (heme) is located at the center of the molecule and is well covered or hidden, being exposed to solvents in a small proportion of 0.06% of the total molecular surface.
From those presented above and from Table 12.2, we can see that NBSs can be classified into two groups according to the biological component.
The protein has a positive potential of +9 mV due to excess lysinic base debris. There is a 324 Debye dipole moment, which produces an imbalance in the spatial distribution of the acidic chain balance. A number of lysine residues are distributed around the solvent to which the center of the heme that interacts with the redox proteins is exposed (Nelsen et al. 1990).
12.3 Integration of Biological Components into NBSs
NBSs are classified in three generations. At the first-generation sensors, the biocatalyst is attached to the surface of the membrane, and then this arrangement is fixed to the surface of the transducer. The adsorption or covalent attachment of the biologically active component to the surface of the transducer allows the elimination of the semipermeable membrane, which is the second generation. The direct linking of the biocatalyst to the electronic device that translates and amplifies the signal, such as the compact transistor, is the basis of the third-generation NBS miniaturization. Depending on the nature of the immobilization and the interaction between the three components, the A–R membrane contact with the electrodes to the transducer, and the processes in the NBSs have evolved over a generation.
First, the specificity and selectivity are dominated by the biological component and are directly related to its nature: enzymes, antibodies, and microorganisms. The specificity derives from the binding of the analyte to the biological component used as the receptor. At the base of this dominant sequence is the A–R biochemical reaction and the collision process between A and R. Second, the transport of the analyte to and through the surface considered to R is also an important factor. This process is related to the transport of a physical size through typical diffusion, migration, and convection mechanisms. Third, the NBS signal is dependent on the A–R reaction assumed to be at a constant speed.
Transient states and biochemical pseudo-equilibrium conditions are dominated by the reaction kinetics, the nature of the transport, which in turn is coupled with the immobilized A–R interface substrate reactions. Even in the case of a real equilibrium, the reaction speed near the steady state will be important in determining the response time. The kinetics of these processes require additional conditions (Agrawal et al. 2012).
The new types of nanoscale materials with different levels of biocompatibility, the new generation of biocompound cells based on a better understanding of metabolism, the manipulation of information stored at the molecular level all have led to a generation of NBSs with a high level of integration. Molecular information initially stored in the base molecular components can be expressed directly to a higher level called “supramolecular” where interactions between molecules are performed by preestablished algorithms, leading to adaptive, functional, and intelligent materials. Materials are built on conceptional, supramolecular, and combinatorial principles. Separation, storage, and detection techniques are developed using “biomimetic” membranes that function according to biological models or precise physicochemical principles (Li et al. 2014).
12.4 NBSs Based on DNA, Nanotubes, and Semiconductor Polymers
Electrochemical NBS with DNA is the result of medical diagnosis requirements to quickly and accurately determine the segments of a DNA sequence. Results from genetics, molecular biology, and nanotechnology have led to one of the most accurate detection methods: electrochemical NBSs with DNA (combining the principle of NBS ISFET with molecular wires from nanotubes). The operating principle consists of collecting the signal between two electrodes—one working electrode and another reference electrode. The auxiliary electrodes have a specified role in their turn. The sensing mechanism consists in modifying the I–V characteristic (current–voltage) in the presence of a target molecule. Carbon nanotubes are exceptional for the work electrode with high electron transfer velocity and excellent spatial resolution. Target in NBS DNA is an unknown sequence of DNA (or oligonucleotides) attached by functionalization to the carboxyl or amino CNT groups. Researchers reported the developing plants’ ability to capture 30% more energy by implanting carbon nanotubes into chloroplasts and plant organisms where photosynthesis takes place. They managed to modify plants so as to detect nitric oxide by implanting another type of carbon nanotubes. “The plants are suitable for the role of a technological platform. They heal themselves, they are durable, they resist the harsh environments and they have their own sources of energy and water.” The transformation of plants to photon devices, with own energy, such as explosive detectors or chemical weapons, is expected (Panchal and Upadhyay 2014).
External or surface electrodes are metal electrodes that contact the NBSs’ bioactive component either directly (dry or solid electrodes) or through an electrolyte solution (liquid electrodes). Solid or dry electrodes are made in silver, platinum, gold, and nickel. Internal electrodes are made of thin wires made of durable metal: stainless steel, platinum, and tungsten. The active part of the electrode can be covered with a metallic conductor layer (gold, silver) and the inactive one with an insulating layer (polymer/thin film). NBSs’ active contact surfaces are large in size compared to cell sizes and are used for extracellular recordings.
Microelectrodes are internal electrodes, but they are built to measure potentials in direct contact with the receiver in NBSs. The contact surface with R is micronized. Microelectrodes can be solid, compounds which can be achieved by depositing a conductive layer (platinum, gold) on a glass support having a particularly thin peak, and another constructive variant consists of inserting a metallic or carbon fiber conductor into an epoxy resin support mixed with a conductive paste; they are used in cellular samples and consist of a glass pipette having a micronized tip filled with an electrolytic solution containing potassium chloride. In the electrolyte solution, a conducting wire is immersed to pick up the electrical potential (Wu et al. 2014).
12.4.1 The Gold Electrode
It was thought that it is not possible to transfer electrons directly between the electrode and the proteins due to their distortion. Several practical considerations have led to the conclusion that the active center of the heme is irreversibly adsorbed when resulting in protein denaturation in contact with the electrode. Changing the surface of the gold electrode by surface adsorption of 4,4′-bipyridyl resulted in the modification of the electrode surface configuration for interaction with cytochrome “c.” 4,4′ Bipyridyl is not an electroactive substance in the potential region and therefore does not play a role as a mediator. This electrochemical addition was possible due to the quasireversible binding of cytochrome “c” to the modified gold electrode with 4.4′ bipyridyl, thereby resulting in the hydrogen linkages in the lysine residues to bound to the nitrogen of the pyridyl which modified the surface of the electrode. Transmutation through complex protein electrode rapidly directs the transfer of electrons, which is accomplished by the following scheme: cytochrome diffusion “c” on the electrode, protein binding on the surface, electron transfer, and protein desorption. Following this process, more than 60 surface changes were possible for electrochemistry of proteins and the gold electrode. Using a bifunctional reagent (X-Y), wherein the X group is the N, P or S bonded electrode and the Y group, which must be bonded “represent also examples of patterns developed” (Agrawal et al. 2012).
12.4.2 Graphite Electrode
Electrochemistry of proteins has been extended to the carbon electrode. Pyrolytic carbon–graphite forms, vitreous carbon and mesocarbon, are structures in which graphene plans are arranged in ABABA hexagonal mesh or disordered in different turbulent forms. The base graphene plan is hydrophobic, but existing or induced defects lead to free C–C bonds, and there is an increase in C–O linkages by oxidation. The direct electrochemistry of positively charged proteins can therefore be performed on the edges of the graphite plans of the carbon electrode. The direct transfer of electrons to negatively charged proteins, such as plastocyanin with graphite electrode (edges or plane edges), can be aided with Mn, Ca, Cr complex cations complexed with amino compounds, Cr (Am6)3+, used as promoters of reactions. In this context, the promoters are inactive redox species in solutions but allow the transfer of electrons to the redox proteins. Microminiaturized electrodes have specific advantages among which we mention the improvement of polarization and contact with biological material at the active sites.
Specificity and selectivity depend on the NBS receptor biological component and its affinity for the analyte. Affinity is a specific feature of enzymes, antibiotics, and receptors being used in functions in living organisms. Affinity is based on the chemical coupling between a component and its complementary partner. In the case of high-affinity components, the diffusion process is rapid, leading to the formation of the Ag–Ab type of complex. The association reaction specific to molecular recognition will be characterized by the first-order response rate constant. In NBS measurements it is essential to consider the concentration of a constant component and other variables. The results of electrochemistry of proteins have been extended to amino acids and peptides (Barroso et al. 2015).
12.4.3 Kinetics of Enzymes Used in NBSs
The addition of enzymes to solutions containing substrate molecules is the essential condition in enzyme catalysis reactions. Extracting the necessary information from the enzyme science to be applied to the development of NBSs such as the enzyme electrode is an extremely difficult task. References will be used to outline some of the enzyme properties necessary to describe enzymatic NBSs. Consider a simple reaction, with a single substrate S, that combines with enzyme E to form the enzyme–substrate intermediate complex, ES.
This unstable complex undergoes a new reaction resulting in product P. The formation and consumption rates of the complex are equal. As soon as E and S enter the reaction, the system becomes unbalanced, the concentration of the complex will be zero, and the formation speed of the complex is much higher than the rate of its consumption. As the reaction unfolds, ES increases and implicitly increases the rate of disappearance of the complex relative to its rate of formation. Initially, the excess of the substrate determines the consumption of the enzyme, and during the course of the reaction, the enzyme’s constant regeneration begins to reach its steady state. The analysis of these reactions results in two important conclusions:
-
At a low concentration of the substrate, the rate of the reaction is proportional to the substrate concentration and inversely proportional to the rate of the formation and extinction rate of the complex or the dissociation reaction rate in the initial reactants plus the decomposition reaction rate in products.
-
At a high concentration of the substrate, maximum speed is limited by enzyme concentration. Thus, the two sequences correspond to the two processes that can control the overall reaction rate (Stein et al. 2011).
When the reaction takes place in homogeneous solutions at a uniform rate, the same in the entire environment, it is necessary to consider the change in the concentration of the components over time. Three mechanisms of mass transport occur in solution: diffusion, convection, and migration.
12.5 Recent Advances in Electrochemical NBSs
An electrochemical NBS is an autonomous, self-contained device capable of providing specific quantitative or semiquantitative analytical information using as a molecular recognition element, a biochemical receptor (biological identification element) that is in direct spatial contact with an electrochemical transducer. Electrochemical NBSs are distinguished only by the nature of the transducer regardless of the nature of the biological component according to the classification in Table 12.3.
Due to their ability to be calibrated in a repetitive manner, an electrochemical NBS is distinguished by a bioanalytical system requiring additional processing steps, such as the addition of reagent. NBSs for a single type of measurement, or unable to continuously monitor concentration analysis or not to be rapidly and reproducibly regenerated, are defined as “disposable.”
NBSs are classified according to their biological specificity—with reference to the mechanism or to the interpretation of the physical–chemical signal (the transducer) (Barroso et al. 2015).
The biological recognition element is based on a catalyzed chemical reaction or an equilibrium reaction with macromolecules that have been previously isolated or synthesized in their original biological environment. In the case of reversible reactions, the steady state can be reached if there is no net consumption by the agent of the immobilized biocomplex and incorporated into the NBSs. Electrochemical NBSs can be classified according to the analyses and reactions they monitor: direct monitoring of the concentration of the analytes or their production or consumption reactions and, alternatively, an indirect monitoring of the inhibitor or activator of the biological recognition element (the biochemical receptor).
Criteria include calibration characteristics (sensitivity, linearity, operational range of concentration, quantitative determination limits, and specific detection), selectivity, equilibrium state and response time, reproducibility, lifetime, and stability (Fang et al. 2015).
The notion of recognition is used in NBSs or in nanobiosensory systems by association with the sensory systems of the plants.
Sensations such as smell or taste are made up of systems that contain an identification receiver cell coupled with neurotransmitter signal-processing pathways. Such phenomena also occur in biochemosensors but at a much-simplified level compared to the complexity of molecular recognition in living systems (Barroso et al. 2015).
Examples of single or multiple transfer signals, limited to the main biochemosensors, are shown in Table 12.3. For the receptor types shown in Table 12.3, different electrodes and measurement methods can be selected from Table 12.4 to form an electrochemical NBS. NBSs are classified by the recognition element (Table 12.3) or by the transduction mode (Table 12.4). NBSs irrespective of the type of classification should be treated unitarily as a microsystem, the biological recognition element being in direct contact with the transducer element.
An electrochemical NBS is an NBS with electrochemical transducer (Table 12.4). It is considered to be a chemically modified electrode (CME), the electric conductor that transmits the electrons from the interaction process to the outside in the electronic measuring system. The electrode may be a metal, a semiconductor, or an ionic conductive material coated with a biochemical or bioactive film. Electrochemical NBS is an integrated transducer microsystem capable of providing selective, quantitative, or semiquantitative analytical information using a biological identification element. It can be used to monitor biological and nonbiological elements. Chemical NBSs that incorporate nonbiological components as receptors, although used to monitor biological processes (pH or NBSs of oxygen), are not NBSs.
The Clark electrode is of importance in the NBSs’ measuring range. Similar physical NBSs used in biological environments such as those measuring pressure, etc. are not considered NBSs (Jacoby et al. 2015).
12.5.1 Classification
Electrochemical NBSs according to the terminology set out in Tables 12.3 and 12.4 can be classified according to their biological specificity, by mechanism or mode of signal transmission, or alternatively, the combination of two.
They can be amperometric, potentiometric, field effect (FET), or NMSs’ conductometric (electrical conduction measurement) respectively impedance metrics. Alternatively, they may be called enzymatic amperometric NBSs to specify the nature of the receptor and the transducer. The first NBSs that were studied are the enzymes and immunosensors (Fang et al. 2015).
12.5.2 The Biocatalytic Recognition Element Receptor
NBSs are based on a catalyzed reaction of biomacromolecules present in the original biological medium that is preisolated or synthetically produced. The reaction is monitored by an integrated detector (transducer) that measures the stationary or transition states or the final reaction product via the immobilized biocidal product in NBSs. Types of commonly used biocatalysts are enzymes (simple or enzymatic complexes)—most commonly used as recognition systems, cells, microorganisms (bacteria, fungi, eukaryotic cells, or yeasts), cellular organs, or component (cell walls, mitochondria) sections of plant or animal tissues.
NBSs with biocatalyst recognition elements are the best known and studied since the beginning of their approach by Clark and Lyons. One or more analytes, commonly called S and S′ substrates, react in the presence of one or more enzymes, cells, etc., to produce the P and P′ products (Fang et al. 2015). There are four strategies whereby the associated transducer can monitor the consumption of S-analyte through the biocatalytic reaction:
-
Detection of S′ cosubstrate consumption, oxygen depletion through the oxidase-induced reaction chain, bacteria, or yeast. The measured signal is the decrease in cosubstrate consumption compared to the initial value.
-
Recycling of the reaction product P such as peroxyhydrogen, H+, CO2, and NH3, in oxidoreductase reduction schemes, hydrolysis, lysis, etc. The signal from the transducer will be amplified.
-
Detection of active centers in the biocatalyst: redox, cofactors, prosthetic groups evolving in the presence of substrate S by using an immobilized mediator. It reacts quickly with the biocatalyst and is easily detected in the transduction chain. Various ferrocene derivatives, such as tetrathiafulvalene, tetracyanochinodimetane (TTF + TCNQ), organic salts, quinones, quinone dyes, Ru, or Os complexes in polymeric matrices, can be used as mediators.
-
Direct electron transfer is made between the enzymatic redox reactive site and the electrochemical transducer. The third strategy eliminates, partially or totally, the dependence of the NBSs’ response on the cosubstrate concentration, S′, which decreases the influence of interference between species.
The use of mediators leads to the decrease of the substrate concentration together with the reaction chains by using a suitable membrane, whose permeability favors the transport of the cosubstrate. When enzymes are immobilized within the same reaction chains, it can improve the performance and abilities of NBSs. Three possibilities are commonly used:
-
Some enzymes facilitate biological identification by sequentially converting the products of the enzyme reaction series into an electroactive final form: this way allows for a wide range of NBS analysis.
-
The enzymatic complex, applied in series, can regenerate the cosubstrate of the first enzyme and amplify the NBS output signal by regenerating another cosubstrate of the first enzyme.
-
The parallel enzyme complex improves selectivity of NBSs by lowering the local concentration of electrochemical interfering substrate: this sequence is an alternative to the use of a permselective membrane or a sequential method (e.g., interpretation of an output signal generated by an NBS and a reference NBS without biorecognition element).
The operation of NBSs is based on the interaction between the analyte and the macromolecules or organized molecular assemblies. Upon reaching the balance, there is no consumption of analyte by the biocomplex agent immobilized in the substrate. The response to the biocomplex analyte–reagent reaction is monitored by an integrator detector. In some cases, the biocomplex reaction is self-monitored by a complementary biocatalytic reaction. The integrator detector monitors stationary or transient states. Antibody–antigen interactions, the most relevant examples of NBSs using biocomplex receptors, are based on immunochemical reactions, e.g., binding an antigen (Ag) to the characteristic antibody (Ab).
Complexes formed by Ab–Ag can be detected provided that other nonspecific reactions are minimized, for each determination of Ag corresponds to a certain Ab that must be isolated, purified, etc. Some recent studies have analyzed direct monitoring of Ag–Ab formation using ion-selective field effect transistors (ISFETs). Increasing the sensitivity of immunological NBSs is achieved by adding specific enzymes to Ag–Ab couples, but this requires additional synthesis steps. As the binding strength or affinity constant varies widely, these systems operate irreversibly (disposable NBSs) or are coupled to flow injection analysis (FIA) systems; then Ab can be regenerated from dissociation of the complex with agents such as glycine–HCl to pH 2.5 (Kurien et al. 2010).
12.5.3 Receptor: Antagonist/Agonist
Recently, they have been used as molecular recognition systems in conductometric analysis, ISFET, or optical NBSs with receptors with ion channels, membranes, or protein structures. A transporting protein, lactose–permease (LP), can be incorporated into a liposomal bilayer that permits protonic carbohydrate transport with a stoichiometric ratio of 1:1. This mechanism was identified through the pH-dependent fluorescence of a fluorophore immobilized in liposomes. Liposomes with LP were incorporated into a lipid bilayer deposited on a pH-sensitive ISFET. Preliminary results show that this modified ISFET is capable of irreversibly detecting lactose from an FIA system.
Protein receptor NBSs have been recently discovered. Binding of analytes, here called agonists, to immobilized receptor proteins is monitored by changing the flow of ions through these channels. Glutamate, as an agonist, can be determined in the presence of other agonists that can interfere with the determination of Na+ or Ca2+ streams using conductometric method or ion-selective electrodes. Due to the dependence of the ionic channel on the nature of the linkages, it produces an independence toward the enzyme nature in order to achieve the desired sensitivity. Two methods have been approached. The first refers to oligonucleotide duplex interleaving during the formation of the double helix structure of the DNA of a molecule that is electrically active. The second method is the direct detection of guanine which is electroactive. Some of these NBSs cannot operate through analytical-sensing membrane separation membranes. The sensitive layer often has to be in contact with the biological environment where the analytes are located (Fang et al. 2015).
12.5.4 Indirect Inhibitor or Activator Monitoring of Biochemical Receptors
NBSs have been developed for indirect monitoring of organic pesticides or inorganic compounds (heavy metals, fluorides, cyanides) that inhibit the biocatalytic properties of enzymes used in the construction of NBSs (devices are irreversible). In immunosensors, the initial biological activity can only be regenerated by chemical treatment and therefore is not part of the reconditioned or reusable NBS class. Their application potential is to warn and not to accurately monitor a specific analyte (considered as disposable). NBSs with cyanide (i.g. inhibition of cytochrome c oxidase) that are used as inhibitor to cytochromoxidase are regenerated by washing with a phosphate buffer at pH 6.3 (Armstrong and Beckett 2011).
12.5.5 Immobilization of Biological Receptors
With the development of enzymatic glucose NBSs, an experiment in which glucose oxidase is immobilized between two membranes, literature has emerged about techniques for immobilizing biological receptors. Enzymes, antibodies, cells, or tissues with high biological activity can be immobilized in a thin film on the surface of a transducer through a variety of methods. The following immobilization procedures of biological receptors are used:
-
Immobilization on the membrane on the surface unexposed to the analyte: an enzymatic solution, a cell or tissue suspension, rests between the permeable membrane to the analyte and the measuring electrode (electrochemical detector).
-
Retaining of biological receptors in a polymeric matrix, polyacrylonitrile, agar gel, polyurethane (PU) or polyvinyl alcohol (APV), redox hydrogels with redox centers such as [Os (bpy) 2Cl]+/2+.
-
Retaining of biological receptors between self-assembled layers (SAM) or in membranes from the double lipid layer (BLM).
-
The covalent binding of membrane surface receptors through bifunctional groups: glutaraldehydes, carbodiimides, SAMs, avidin–biotin silanized.
-
Modification of the entire electrode structure (modified carbon paste with enzymes or graphite in epoxy resins).
Receptors are modified either alone or mixed with other proteins, such as bovine serum albumin (BSA ), either directly on the surface of the transducer or in the polymer membrane. Reactivated membranes can be used directly to immobilize enzymes or antibodies without chemical modification. The covalent binding and cross-linking are more difficult than immobilization or the retaining of receptors on the membrane. In the case of microsensor structures where the membrane is directly deposited on the transducer, the covalent bonding is safer and more stable (Muñoz et al. 2008).
12.6 Internal Membranes and External Membranes
Besides reactive layers or membranes with immobilized receptors, many NBSs, those for clinical or biological applications, incorporate one or more internal or external separation auxiliary membranes with three important functions: the barrier, the outer diffusion barrier for the substrate, and the biocompatible surfaces. For any NBS built on the principle of molecular recognition, it is important to characterize it by its response, which is related to operating parameters and limiting reaction speeds. Accuracy, precision, sensitivity, and reproducibility are basic criteria for estimating NBS performance. These parameters are in direct relation to the reaction mechanisms, the transport phenomena, and the kinetics of the processes in the volume at the interface. Most criteria have been developed for enzymatic NBSs, being the most studied in the literature. In the case of immunosensors, the key element is the ability to capture the surface, i.e., the number of surface molecules that is active. One method of checking this parameter is to measure the specific activity, meaning the ratio of the number of active molecules to the number of immobilized molecules. This estimation is dependent on the immobilization mode (molecular orientation, number of attachment points or active sites), and the ratio ranges between 0.15 and 0.3, rarely reaching the unit. Capture capability becomes important when the surface decreases as in microfluidic applications. A problem encountered in immunosensors is that of regenerating the surface without significant loss of activity. There was a lack of rigor in the performance criteria (Affi et al. 2016).
The response signal is corrected for background noise, the reference concentration is usually estimated in mol/L although this high value is never used when measuring ranges refer to 10−10 mol/L, and currently sensitivities of the order nmol/L and pmol/L have been reached.
Transient response is important for dynamic assay analysis and sampling techniques but is less significant in continuous monitoring. The transient response is estimated by the slope (dR/dt) max after the addition of the analyte in the measuring cell. One evaluation method is to introduce NBSs into an FIA system for sequential sample analysis in a specified hydrodynamic regime. The sensitivity and linear range of measurement of stationary concentration are determined through graphical representation. This method is more concise than the current calibration curves used to plot the response corrected to the baseline based on its concentration or logarithm. Parameters are estimated in the linear response range of NBSs.
Any electrochemical NBS has a superior linearity of the response. This limit is directly related to the biocatalytic or biocomplex properties of the biochemical or biological receptor. More in the case of NBSs with enzymes, this limit is significantly influenced by membranes and immobilized substrates where the diffusion barriers and secondary kinetics play a role. The local concentration of the analyte in the reaction layer may be two orders of magnitude smaller than the volume of the solution (Michelini and Roda 2012).
Enzymatic kinetics are described by Michaelis–Menten mechanisms and expressed by KM and VM parameters. For the kinetics of the enzyme in the solution phase, KM is usually determined from the Lineweaver–Burk graphical representation. For any electrochemical NBSs, the number of standards used and how the standard sample matrix can be simulated or duplicated should be set, being required to specify the procedures for each type of NBSs related to its application. These are important for the disposable NBSs’ case using immunoaffinity or inhibition reactions. Sensitivity is the slope of the curve and should not be confused with the quantified detection limit (LOD) relative to the baseline or noise signals. The range of work concentrations is determined by lower or higher detection limits (Fang et al. 2015).
Selectivity and safety are determined as any kind of amperometric or potentiometric NBSs. They depend on the choice of receiver and transducer. Most enzymes are specific, but there are also nonselective enzyme classes, such as alcohol oxidases, the group of oxidases sugars, peroxidases, lactases, tyrosinases, ceruloplasmin, alcohol dehydrogenases, glucose dehydrogenases, NADH dehydrogenases, etc.
They have been used to develop NBSs to determine environmental phenols or to monitor food quality. On the other hand, oxygen electrodes, pH electrodes, and ISFETs have a pronounced selectivity, the same as metal electrodes that are sensitive to many substances. Their selectivity may be changed when these transducers are associated with receptors. ENFET is pH sensitive to the buffer and protonation but its selectivity is not altered. When the transducer interferes with other substances, known as ascorbate or urease, to glucose NBSs based on hydrogen peroxide detection, these side effects may be restricted by the use of outer or inner permselective membranes.
Alternatively, NBSs with and without biological receptors that work by differential NBSs are designed. Safety in operation of NBSs depends on the selectivity and reproducibility and accuracy of the measurements (Heyl et al. 2012).
12.6.1 Clark Electrode: Applications in Plant Nanobionics
The Clark construction principle studied electrochemically oxygen as a reducing gas and platinum as a metal electrode. Platinum used for detecting electrochemical oxygen is known as the Clark electrode. The electrode has an organic membrane covering the electrolyte layer and two metal electrodes. Oxygen diffuses through the membrane and is electrochemically reduced to the cathode. Between the cathode and the anode, a fixed voltage is applied, for which the oxygen reduction reaction takes place.
Temperature greatly influences reaction speed and solubility. This is a polarographic electrode used to measure the concentration of oxygen in body fluids or gases. The sample is in contact with a membrane (polypropylene or Teflon) through which the oxygen diffuses into a measuring chamber containing 50% saturated potassium chloride solution. Inside the room are two electrodes, one is reference, Ag/AgCl, and the other is platinum, coated in the glass.
The electrical current at the polarization potential of –600 mV is proportional to the oxygen concentration in the solution. For reverse polarization at +600 mV, hydrogen measurements can be made. Reactions are very sensitive to temperature and should be maintained at ±0.1 °C. The electrode is calibrated using a mixture of the two gas–oxygen and hydrogen-known concentrations. Oxygen electrode or Clark electrode has proven to be an analyzer of raw gas or developed gas when performing chemistry analyses in the clinical laboratory and in the field of medical care, ambulatory, or intensive care (on the surface of the platinum electrode an enzyme reacts with oxygen).
The enzymes are placed in a closed membrane to the surface, which can be recognized as the simplest model of NBSs. The oxygen concentration curve was proportional to the glucose concentration. It was the first NBS built, which helped the progress of laboratory analyses a lot. Oxygen diffuses through the membrane and is electrolytically reduced to the cathode. The higher the partial oxygen pressure, the more oxygen diffuses at a time.
The temperature NBSs attached to the sample allow the membrane to compensate for the diffusion and solubility rate. The measuring instruments record cathode current, sample temperature, membrane temperature, barometric pressure, and salinity. With this information one can calculate the oxygen contained in the sample, either in parts per million (ppm) or in percent of oxygen saturation. The geometric configuration of the Clark electrode is of great importance.
In particular, the thickness of the electrolytic layer between the cathode and the membrane must have a certain limit, to ensure linearity and decrease of the drift current. Calibrating a polarographic system is a must. Proportionality between current and oxygen concentration must be ensured, with errors below 1% (biological samples role and air parameters are essential).
Air, as a gas mixture that has a constant oxygen content of about 20.9%, when in contact with water, the dissolved amount depends on several factors: the optimal time for oxygen dissolution, homogeneity of the water solution, water temperature, air pressure, salts contained in water, and other water-soluble substances that are oxygen-consuming. Oxygen contained in water is determinant for biological and chemical processes, so measurement of dissolved oxygen in water is important to find the partial pressure of dissolved oxygen; it must be saturated in pure water at a certain temperature (Wolfbeis 2015).
12.6.2 The Enzymatic Electrode
The enzymatic electrode (in some references known as the enzyme electrode) is a combination of an electrochemical probe of any type (amperometric, potentiometric, or conductometric) with a thin layer (10–200 microns) of immobilized enzyme. In these devices the function of the enzyme is to provide selectivity in virtue of its biological affinity for a substrate of molecules. An enzyme can catalyze a reaction of a given substrate for a specific isomer from a plurality of substrates with different isomers.
Typically, the degree of advancement of an enzymatic reaction (directly related to the concentration of the analyte) is monitored by the rate of product formation or the disappearance of a reagent. If the product or reagent is electrically active, then the response can be directly monitored by amperometry, i.e., the variation of the current for a given applied potential.
The main considerations are: Does the enzyme contain active redox groups? Are the biochemical reaction products electroactive? Is one of the substrates or cofactors electrically active? What is the speed and response time? What is the final application of NBSs? If the enzyme does not contain redox groups, then NBSs are limited to measuring the product or substrate consumption by their reaction to the transduction electrode. The electrical current is directly related to the analyte concentration. NBSs are based on electrochemical response due to H2O2. Most common enzymes used in the design of enzyme electrodes are those that contain redox groups that change their redox state during the biochemical reaction. Redox enzymes are oxidases and dehydrogenases, pyrroloquinoline quinone (PQQ). They act by oxidizing the substrate, accepting electrons during the process, and further transforming in a reduced state.
These enzymes return to the oxidized active state by transferring electrons to the oxygen molecule resulting in hydrogen peroxide (H2O2). Both oxygen and peroxide being electrochemically active, they continue by reducing to cosubstrate (oxygen) or oxidation of peroxide (reaction product).
The method based on the reduction of oxygen to the O2 electrode is one of the simplest methods but suffers from several disadvantages, namely, slow response, miniaturization difficulties, low accuracy, and reproducibility. Measurements on peroxide oxidation overcome the above difficulties and are currently the most popular method. Mediator systems—a major limitation of the above—described hydrogen permeation system is the high operating potential (about 0.8 V against the Ag/AgCl reference electrode) required for oxidation of peroxide which leads to increased interference. The use of mediators (molecules that can carry electrons between the enzyme redox center and the electrode) can minimize this inconvenience.
Depending on the nature of the mediators, the potential applied may be reduced below the limit of interferences of species such as ascorbate, urate, and paracetamol. A large number of compounds are able to act as mediators in the enzyme electrode. Of these, the most popular are the metallic complexes. Representatives for organometallic complexes are ferrocenes and their derivatives because they have redox potentials and are independent of pH. Bienzymatic systems’ recent works have focused on the direct communication of electron transfer between enzymes and electrodes. Successes in the field are the peroxidase enzyme HRP (horseradish peroxidase) that catalyzes the reduction of hydrogen peroxide for a number of organic compounds. When the enzyme is immobilized on the electrode, the need for the organic reducer is prevented by the electrode itself providing the reducing equivalences. The coupling of peroxidase with an enzyme oxidase allows for the construction of bienzymatic systems through which the peroxide produced by the oxidase is detected by the electrode–peroxidase system which operates at lower potentials relative to a simple platinum electrode. This is where the minimization of active species interferences results from (Frederickson Matika and Loake 2014).
12.7 Fiber-Optic Biosensors (FOBS) in Plant Nanobionics
Optical fiber as a nanobiosensor can be placed in the surface or inside the plant to directly measure parameters. NBSs with optical fiber are proposed to be used in many and rapid medical determinations, and its applications are continuously expanding. It can be attached inside a hollow-like tubular instrument, serving to dilate a hole or channel, and inserted into the tissue, performing a minimal monitoring where it is needed. NBSs with optical fiber are nontoxic, chemically inert, and can be successfully used inside the plant. It can be associated with plant monitoring equipment. It’s easy to handle with negligible weight.
The evolution of fiber-optic NBSs is based on multiple performance and biocompatibility. Biocompatibility is the first step in the plant’s comfort; NBSs should not affect the physiological parameters of the plant, but its functionality must not be compromised by plant disassimilation products. Fiber-optic NBSs can be classified as extrinsic, fiber acts as a way for signal and intrinsic, interactions occur in the fiber itself. There are two types of FOBS: minimally invasive NBSs that are introduced into the cavities of the plants and invasive NBSs that are introduced into the organs or in wood conductive tissue (Liu et al. 2015).
12.8 Applications in Plant Nanobionics
In the last decade, optical fiber is a product that is widely used in all the cutting-edge fields of advanced science and technology. Given the ease with which it can be manipulated, unlimited sterilization possibilities, and reduced costs, it can be estimated that this product will increasingly gain market. The following applications are known to have used fiber-optic NBSs: in epidermis and vascular tissues, for analysis of raw sebaceous elements, saturation in oxygen, raw sewage gas analysis, sap pH; in plant breeding monitoring; easy pH determination with a microabsorbent indicator and pH–modulator, acid–alkaline; in vegetal tissues, when it is intended to monitor the temperature, or to diagnose small and very small injuries that are difficult to reach; in epidermis for can test the quality and integrity of the layers, so, small lesions can be detected, can be used to stimulate tissues, a FOBS based on oxygen demand (BOD) can be used; in the stem can identify very small injuries that are inappropriate. Another possible application is to appreciate the color or integrity of the vascular tissues. Optic-fiber NBSs can now monitor electrolytes from raw or elaborated sap as well potassium, sodium, and calcium. It takes the form of a tubular instrument, able to expand an optic-fiber channel (0.5 mm diameter) that can be inserted into vascular tissues. It can measure the gas concentration and the pH of the raw or elaborated sap and oxygen saturation also.
The materials that make up the chemical transducers are ionophores that can be reversibly attached to the electrolyte by a molecular separator (spacer) and fluorophore, respectively. The degree of fluorescence, through excitation with electroluminescent diodes in purple, is modulated by ionophores proportional to the analyte concentration. NBSs are used either extracorporeally in the external raw or elaborated sap gas analysis circuit or intracorporeally for continuous blood gas monitoring in critical situations. The chemical parameter capable of monitoring cell state is pH because lactic acid, formed when cell tissue dies, produces a decrease in pH. Any drop in the pH of the raw or elaborated sap from 7.4 indicates cell death (McLamore et al. 2010).
Achievements in the domain are invasive pH NBSs that determine the state of the cell. NBSs are composed of a fluorescent dye encapsulated in a gel matrix (polyacrylamide) attached at the end of the optical fiber. The dye is characterized by the emission of the acidic form centered on 580 nm and the alkaline form centered on 680 nm. The two forms are pH sensitive.
Excitation occurs at 533 nm for both forms. Separation of emission is done directly through optical filters, and sensitivity is 0.05 pH units far below acceptable clinical standards (0.1 pH). A bacterial disorder is a multifactorial condition that is characterized by demineralization of the inorganic portion and an organic destruction of the substance. Each bacterial perturbation has as an etiological agent as pathogenic species. The content of the raw or elaborated sap in terms of bacterial load is about 109/mL raw or elaborated sap. Therefore, raw or elaborated sap can be considered as a selective medium for bacterial growth. A significant correlation has been demonstrated between the number of pathogens in the raw or elaborated sap and their prevalence in the problems. A simple method has been adapted for detecting and counting the pathogen; it is a device consisting of a special support made with an agar culture medium containing 20% sucrose. It is inoculated with raw or elaborated sap, and the density of growth of the pathogen is assessed after incubation for 48 hours at 37 °C. Next comes the morphological identification of distinct colonies on selective and nonselective agar, on distinct cell form, visible in the light of the microscope.
The technique also has some disadvantages that it takes time for bacterial growth and also requires additional laboratories. To monitor pathogen activity in the sap, a FOBS was developed that monitors the pathogen-mediated sucrose reactions through a photosensitive indicator immobilized in a porous glass coating. The surface of the optical fiber core is treated or coated in a porous glass film using the sol–gel technique (Kozan et al. 2007).
Spectroscopic analysis showed that there are two phases in light absorption at 597 nm over a duration of 120 min: between 0–60 min and 60–120 min. The investigation shows the potential of NBSs in monitoring plant activity. The sol–gel technique is used to immobilize the photosensitive indicator, and it is simpler than compared with the principle of selective medium which takes time and is laborious. Criteria at the base of the experiment: pathogens are partially anaerobic with optimal growth at 37 °C; glucosyltransferases and fructosyltransferases from pathogens catalyze the synthesis of water-insoluble glucan and fructanic polymers in sucrose to form lactic acid found in acidic sap; pathogenic agents in the sap synthesize both extracellular and intracellular polysaccharides from sucrose; extracellular polysaccharides help the adhesion of bacteria to the surface, while intracellular polysaccharides are stored for bacterial energy; polysaccharides, intracellularly, help the bacterium to continue fermentation even when there is no exogenous form of food. Acid tolerance of pathogens causes their activity to continue even at a low pH; a pH indicator, photosensitive, produces a characteristic color, according to a color gradient, depending on the pH of the raw or elaborate sap and is used in the FOBS construction (Miranda et al. 2011).
On the basis of these considerations, an experimental assembly was designed and performed with a double beam UV spectrometer in which optical fiber was introduced instead of a cuvette. In the reference well, the blue bromophenol buffer solution was used for different pH values, from 4 to 7. The initial experiment helped determine the wavelength and peak characteristic to the buffer indicator and for different pH values in the sap, induced by bacterial activity. UV spectroscopic analysis at a pH of 4 and 7 of the blue bromophenol solution showed slightly prominent at 590 nm wavelength; peak intensity decreases from pH 7 to 4. Comparison with literature data showed a good concordance, observing that in the medium with sap, sucrose and bromophenol blue, the absorption was stable at 590 nm wavelength for a time interval of 15 min, 30 min, 1 h, and 24 h. For each set of corroded and processed fibers set, it requires independent calibration due to the in homogeneities resulting from the optic-fiber preparation steps. FOBS proves to be a rapid quantity measurement test of pathogen activity in raw or elaborated sap. This test can also be adapted to other plant nanobionics where bacterial activity is involved in cellular or tissue destruction.
The method of forming a biosensor from an optical fiber for the observation and detection of the pathogen, the experiments of this study followed the phase of biochemical recognition of the signal and the phase of the spectroscopic analysis. The idea of nanobionic plants has evolved to create solar cells that heal themselves from plant cells. The next step was the desire to try to amplify photosynthesis in isolated chloroplasts in plants to be used in solar cells.
Chloroplasts host everything they need for two-step photosynthesis. In the first step, pigments such as chlorophyll absorb light, which generates the stimulation of electrons that circulate through the chloroplast tilacloids. The plant captures this electrical energy and uses it to fuel the second stage of photosynthesis, creating glucose. Chloroplasts also have these reactions after they have been removed from the plants, but after a few hours, they break down because light and oxygen destroy their photosynthetic proteins. Normally, unlike extracted chloroplasts, plants can repair this damaging process. To prolong chloroplast productivity, researchers have attached ceric oxide nanoparticles. These particles are, in fact, powerful antioxidants that remove oxygen radicals and other high reactivity molecules produced by light and oxygen, protecting the decomposition chloroplasts.
The researchers introduced nanoparticles into chloroplasts using a new technique called LEEP (lipid exchange envelope penetration). By wrapping the particles into polyacrylic acid, a heavily charged molecule allowed the particles to penetrate the lipid hydrophobic membranes surrounding the chloroplasts. In these chloroplasts, the level of decomposition of molecules has decreased tremendously. Using the same technique, the researchers introduced semiconductor carbon nanotubes embedded in negatively charged DNA into chloroplasts. Plants use only one tenth of the available sunlight, but carbon nanotubes have functioned as artificial antennas that have allowed chloroplasts to capture unusual light waves such as green, ultraviolet, and near infrared. When carbon nanotubes functioned as prosthetic photoabsorbents, photosynthesis, measured by the activity of electrons in tilacloids, was 49% more intense than in chloroplasts isolated without attached nanotubes. When cerium oxide was joined with carbon nanotubes, the chloroplasts remained active for the next few hours (Nikolelis et al. 2008). Researchers went to live plants and used a technique called vascular infusion to attach nanoparticles to Arabidopsis thaliana, a small flower plant. Using the above method, the researchers applied a nanoparticle solution on the lower side of the leaf, penetrating the stomata that usually allow the carbon dioxide to get in and the oxygen out. In these plants, the nanotubes have penetrated into chloroplasts and have increased the photonic electron circuit by about 30%. It is also to be discovered how these percentages influence the sugar production of plants. Scientists have been able to transform the Arabidopsis thaliana plant into a chemical NBS by implanting nanotubes that detect nitric oxide, a pollutant produced by combustion (Hines et al. 2015).
NBSs have been created on the basis of carbon nanotubes for several chemicals, including hydrogen peroxide, trinitrotoluene, and sarin gas. When molecules attach to polymers encased in nanotubes, the fluorescence of these nanotubes is altered. Carbon nanotubes can be used to create NBSs that detect real-time particle free radicals or signal the presence of molecules with a very low level of concentration and too difficult to detect. This is a tremendous demonstration of how nanotechnology can be combined with synthetic biology to modify and enhance the functions of living organisms of plants. The way that nanoparticles arrange themselves can be used to enhance plant photosynthetic capacity, being used as NBSs and stress reducers.
By adapting NBSs to targets, researchers hope to develop plants that could be used to monitor environmental pollution, pesticides, fungal infections, or exposure to bacterial toxins. Attempts to incorporate electron nanomaterials into plants, such as graphene, are currently being made. Researchers have long tried to find the best way to transmit information in a timely manner, focusing on electronics and mechanics of NBSs for their tasks. NBSs used for agricultural purposes are not new. Recombinant DNA technology now offers the possibility of obtaining new biological insecticides that preserve the benefits of “classic” biological control agents, plus some new features. These technologies are not accessible to all possible users, especially if they are poor, and furthermore, they have also generated a series of public debates about their usefulness and effects on organisms other than the target or the environment.
Obtaining pest-resistant plants is perhaps the most spectacular field of genetic engineering applied to plants, since it allowed the regeneration of plants containing genes of bacterial origin that provide protection against harmful insects. This ensures, on the one hand, the obtaining of richer harvests and, on the other hand, the reduction of farmers’ costs for pesticides (Lukács et al. 2006; Prasad et al. 2014, 2017).
A series of new genes for resistance to insect attack, transferable to plants (genes encoding δ-endotoxin production from B. thuringiensis) has been discovered; genes for the synthesis of enzymes or enzyme inhibitors; plant genes encoding the synthesis of specific lectins; genes that cause induction of synthesis of plant compounds such as phytoalexins.
The development of cloning methodology was based on the observation that there is a group of Gram-positive bacteria belonging to B. thuringiensis species that produce a toxin called δ-endotoxin or crystalline protein capable of killing a wide range of insects (Coleoptera, Lepidoptera, Diptera), depending on the bacterial strain. Of great interest is strain B. thuringiensis var. tenebrionis that synthesizes an effective δ-endotoxin against the Colorado beetle. The genes involved in the synthesis of this protein are localized, on most bacterial strains, to large plasmids (75 kb), the production of the toxin occurring during sporulation. It has been shown that crystalline protein (δ-endotoxin) is normally expressed as a large inactive protoxin, which undergoes proteolytic processing in the intestine of the sensitive insect, becoming an active toxin. It recognizes the specific receptors in the intestinal cells and blocks the functions of these cells, leading to the death of the insects.
Studies on genes that encode inhibitory proteins produced by B. thuringiensis led to their grouping into four types based on target species specificity and nucleotide sequence: type I cry genes, encode specific 130 kDa proteins for lepidopteran larvae; type II cry genes, encode active 70 kDa proteins on dipterous and lepidopteran larvae; type III cry genes, encode 70 kDa specific activity on coleopteran larvae; and type IV cry genes, encode inhibitory proteins for Diptera larvae.
The number of genes identified that are coding for δ-endotoxin-like proteins is high: 140 genes specific for lepidopteran, coleopteran, and diphtheria (Genfa et al. 2005).
It has been achieved to obtain crystalline protein genes from several strains of B. thuringiensis by genetic amplification (PCR). Because the whole gene for the crystal protein was found to be very poor in the transformed plant cells, a modified gene was created, containing only the N-terminal portion of the protein (amino acids 1–645). To increase gene expression in plants, the natural sequence for amino acids 1–415 rich in AT was replaced by a synthetic sequence, rich in GC, containing the preferred codons for plant cells.
These recombinant genes were introduced into Ti plasmid-derived vectors (binary vectors containing the duplicate CaMV promoter, which increases the transcriptional and fivefold transcription and selection marker genes for antibiotic resistance or phosphinothricin herbicide) transferred to cells by A. tumefaciens containing Ti disarmed plasmids. The size of recombinant plasmids obtained ranges between 5000 and 10,000 pb, depending on the size of the bacterial gene and the promoter sequence integrated into the vector. Recombinant bacterial strains were then used to infect test plants (potato, tobacco, cotton). Selection was first performed according to vector-borne selection markers (antibiotic resistance, GUS test, herbicide resistance, etc.), and finally regenerated plants were subjected to insect attack (Takakusagi et al. 2013).
Regenerated plants have shown resistance to attack by insect pests, the specific character being maintained and expressed in experiments in the field. The first transgenic plant that manages the insect attack resistance belongs to the Nicotiana tabacum species, expressing the whole or truncated cry 1A gene, cloned under the control of a constitutive promoter, so that the inhibitory protein represents 0.02% of the total plant protein (leaf). There were obtained cotton plants into which the modified cry 1A (c) gene, cloned under the control of the CaMV 35S promoter or under the control of a promoter and a sequence for a chloroplast transit peptide isolated from Arabidopsis so that the expression level of the gene of interest led to a high level of toxin: 0.1% of total protein, 1%, respectively.
Another variant of cloning the bacterial toxin gene was that of using genetic elements that ensure expression of the gene of interest exclusively in the green portions of the plant (the promoter derived from the gene for PEPC) or pollen (by using a gene derived promoter for a calcium dependent protein kinase (CDPK ). A similar methodology has been used to transform rice plants, and by cloning a synthetic cry III gene, they have obtained tobacco and potato plants resistant to the attack of Colorado beetle (Vigneux et al. 2007).
A modified 1A (b) modified gene was used for cloning under the control of the CaMV promoter and obtained sugarcane plants with resistance to Diatraea saccharalis larvae. Given the practical significance of plant resistance to harmful insects, research has been extended to other plant species, producing eggplant plants resistant to the attack of Coleoptera, broccoli with resistance to certain lepidopteran species, maize with resistance to B. fusca, etc., as well as a number of advances in leguminous plants.
Toxicity studies conducted on plants expressing the gene for δ-endotoxin have shown that the existence of the transgene does not alter the normal features of the plants, except resistance to insect attack. In addition to δ-endotoxin produced by B. thuringiensis strains, other bacterial species have also been identified that produce insecticidal proteins (Liu et al. 2011). This is the case for B. cereus strains producing two VIP1 and VIP2 proteins with effect on insects, their mode of action being similar to δ-endotoxin.
12.9 Genes for the Synthesis of Enzymes or Enzyme Inhibitors with Insecticidal Effect
Expression by plants of enzymes such as chitinase, cholesterol oxidase, lipoxygenase, phenol oxidase, peroxidase, or isopentenyl transferase (ipt) could be an alternative to using the δ-endotoxin gene. Of the enzymes that can provide plant protection against insect attack, a particular place is occupied by chitinases, enzymes that act on chitin, a basic component of insect coatings. Tobacco plants expressing genes for chitinase isolated from insects or beans exhibit increased resistance to lepidopterans. It has been observed that by cloning the isolated chitinase gene from the S. marcescens bacterium, a synergistic effect of endocytinase produced by plants containing the endotoxin gene in addition to S. littoralis larvae has been revealed. Another enzyme of bacterial origin that exhibits insecticidal action is cholesterol oxidase.
The introduction and expression of cho A gene for cholesterol oxidase from Streptomyces sp. in tobacco plants have led to increased plant resistance to A. grandis larvae. The use for cloning the gene for the enzyme bacterial isopentenyl transferase (ipt) (involved in cytokine biosynthesis) by fusion with the protease inhibitor II (PI–IIK) gene promoter determined the production of N. plumbaginifolia lepidopteran-resistant plants (M. sexta or M. persicae).
Another embodiment was that of introducing into the plant cells the cpt2 gene that encodes a trypsin inhibitory protein isolated from Vicia faba. This protein has antimetabolic effect, protecting plants from the attack of pests. Similarly, other genes encoding protease inhibitors (Kunitz trypsin inhibitor, Bowman–Birk proteinase inhibitor) or lectins have been cloned into different hosts, and encoded proteins may be true “defense guns” for the plants that contain them. It is known that insects, such as lepidopterans, depend on serine proteases (trypsin, chymotrypsin, or elastase), these being the first enzymes to digest (Alvarez-Fernandez 2010).
A series of genes encoding different inhibitors for serine proteases have been isolated from various sources (plants, microorganisms) and cloned into various plant species, such as M. sativa, tobacco, corn, etc.; the plants obtained showed increased resistance to various insect pests compared to normal non-GM plants. In some cases, it has been noted that the insertion of additional genes to plants for protease inhibitors to join endogenous genes causes an increased level of pathogen resistance of transformed plants. However, the use of protease inhibitors for controlling insect pests requires thorough studies of plant and insect interactions because it has been observed that for some inhibitors such as serine proteases, the insecticide effect also manifests on useful insects (e.g., on bees). Insect carbohydrate metabolism is another target for inhibitory agents tested in numerous studies. Genes for different α-amylase inhibitors (wheat and beans) were isolated and characterized; after cloning the isolated gene from wheat in tobacco plants, an increase in lepidopteran larval mortality of up to 40% was observed. In the case of cloning the α-amylase inhibitor gene from beans in pea plants under the control of the pha 1 gene promoter, an increased expression of the foreign gene in the seeds was obtained, which resulted in a higher resistance to Callosobruchus sp. (Ramirez and Spears 2014).
Vegetable lectins are a special group of glycoproteins that have protective functions against a number of harmful organisms. Studies on these glycoproteins have shown that they produce strong effects on the development of various types of insects. The first example of plants containing genes for nonspecific lectins that show toxicity to pests is tobacco plants in which the lectin genes from the pea have been cloned. However, many of the vegetable lectins also have a toxic effect on mammals, which limits their use as agents to increase pest resistance. Special attention has been given by many specialists to lectin specific for mangosteen from guinea pigs and concanavalin A: the genes for these lectins have been cloned into different plant species (tobacco, tomato, potato, sugarcane, rice, wheat), and the heterologous proteins synthesized by them have determined a reduced sensitivity to the attack of harmful insects (lepidopteran, aphids, coleopteran larvae).
The results suggest that the use of plants containing insecticide genes (such as for lecithin) together with integrated management represents promising possibilities for controlling pests from many plant species, including wheat or rice (Richard et al. 2014). Contrary to the remarkable results achieved so far, the genes used to transform crop plants are either too specific or only partially effective on the targets of insect pests. To use plants as true “weapons” for pest control, it would be necessary to have genes at their level to determine the synthesis of compounds with different actions on the same target.
The researchers are relatively recent and aim mainly to combine genes for a B. thuringiensis δ-endotoxin with another inhibitory gene in the same host gene: for example, the gene for the V. faba trypsin inhibitor (CpTI) or for serine proteases and the protein gene in the potato virus Y coating. Another interesting approach is that which introduced the cry1A (c) gene into a P. fluorescens strain able to colonize the sugarcane by means of two plasmids, pDER405 and pKT240, in which the gene is found in 13 and 28 children, respectively. Testing of recombinant bacterial strains on sugarcane-specific insect pests revealed greater resistance of plants treated with the respective bacteria than untreated. Also, although pest-resistant plants have been obtained for several plant species, fewer results have been reported for cereals, vegetables, and oleaginous plants (Ibrahim et al. 2008).
Plant resistance to various diseases (microbial, viral, and nematode phytopathogens) has been a subject for long-term studies, identifying a relatively large number of resistance genes. Although it was thought that endogenous resistance genes would provide a sustainable effect for the appropriate plants, this was true in very few cases. In the case of potato, the control program for certain diseases, such as the rot caused by Phytophthora infestans, had to be abandoned because the resistance to this disease of potato plants obtained by transferring the 11 resistance genes on the basis of crossbreeds with the Solanum demissum species proved to be of short duration. Identifying genes of resistance in the genome of different plant species and transferring them to other crop plants are extremely difficult and time-consuming if traditional methods (intra- and interspecific hybridizations) are used. The process is considerably accelerated by the use of molecular markers generated by restriction fragment length polymorphism (RFLP ) techniques, randomized amplified polymorphic DNA (RAPD ), single sequence repeat (SSR ), or single nucleotide polymorphism (SNP).
The application of molecular markers allowed the isolation of nearly 20 resistance genes (R-genes) from genetically well-characterized plant species, proving that many of them are grouped into specific chromosomal regions (they form clusters). Of these resistance genes, some have been transferred to other plant species than to their origin through molecular cloning techniques, with the help of specific vectors that ensure the transfer of large fragments, revealing that in this way, the R-genes act synergistically and provide lasting resistance to some diseases. As it has already been mentioned, few R-genes have been shown to provide a lasting control of plant diseases. This is the case for pepper Bs2 genes and rice Xa21 which provide resistance to different phytopathogenic strains of X. campestris or X. oryzae in the case of species in which the genes have been cloned (e.g., tomatoes). Resistance to these genes is due to the recognition of proteins produced by bacteria or phytopathogenic fungi, resulting in the occurrence of a plant hypersensitivity phenomenon and necrosis of affected tissues. Another example of the long-acting R-gene is the barley recessive mlo gene which provides the resistance of the plants containing it to all E. graminis strains, through the accumulation in plant cells of a phenolic antifungal compound named p-coumaroylhydroxyagmatine.
It is expected that this gene will be used for suppressing the antisense technique of the dominant mlo gene from wheat or other plant species susceptible to Erysiphe sp. In vitro processing of the R-genes and then introducing them into new hosts provide new possibilities for resistance. This is the case for the tomato avrPto gene which, after cloning under the control of a strong promoter such as 35S to CaMV, determines the resistance of transformed tomato plants to P. syringae pv. strains, tomato, and to unrelated pathogens such as X. campestris and C. fulvum. Researchers’ efforts to obtain resistance systems applicable to a larger number of plant species are not limited to R-genes but also include systemic acquired resistance mechanisms (SAR). Several genes involved in SAR have been isolated and characterized, of which the Npr1 gene encoding a transcriptional regulator is a key gene in this system. Overexpression of this gene increases the resistance of Arabidopsis thaliana or rice plants to a wide variety of pathogens (Knecht et al. 2010).
An interesting behavior has been observed in the Myb1 gene which is induced by VMT infection of resistant tobacco plants and which causes the synthesis of a transcription factor that binds to the promoter of a gene involved in pathogenesis (the PR1a gene). Modifying the expression level of the Myb1 gene in tobacco plants leads to increased resistance to viral infection (with VMT) as well as to R. solani pathogenic fungus (Raymond et al. 2007).
Along with this, another recently cloned gene, Pad4, isolated from Arabidopsis thaliana, proves to be extremely interesting for the development of broad-spectrum resistance: overexpression of the gene in plants increases resistance to phytopathogens. Numerous biotech companies and universities have begun to assess the performance of plants that express antifungal proteins through both “in vitro” and field experiments. Both plants containing R-genes or genes involved in SARs, as well as genes such as those which encode the glucosidase (AGO) from Aspergillus sp., defensins, H2O2-generating enzymes, glucanases or chitinases, have been examined. Although at the laboratory level potato plants containing the fungal gene for glucose oxidase showed an increased resistance to phytopathogens, the results were inconclusive when they were put into the field. For other genes such as chitinase and intracellular α-1,3-glucanase, overexpression of these in tomato plants resulted in significant resistance to Fusarium oxysporum (Werner et al. 2016).
Companies have created NBSs that farmers can use to detect information such as air pollution, soil humidity, and so on. However, given that plants are really good NBSs and that they can naturally react to external stimuli and changes, they can be used instead. This is the idea behind the latest NBSs initiative called Advanced Plant Technologies. The idea is that, through genetic manipulation, researchers will be able to create self-sustaining plants, which in turn enable them to act as a kind of NBS when it comes to detecting chemical substances, pathogens, etc. This is not the first time this idea with plants as NBSs appears, but before that, resources that plants needed to survive were used, which in turn reduced their resistance. This new idea indicates that NBSs can be sustained by themselves, which means they can work longer in isolated parts. In the future, plants could be used to detect when a biological attack will take place. In addition, because they are plants, it also means that they can be placed anywhere and nobody will think twice about their presence and that they might be some NBSs.
12.10 Conclusions and Remarks
Through such examples, nature teaches us to optimize by exploring diversity. In this context, integrated nanoscale systems (NBSs), energy sources (biocombustible cells ) that use plants metabolism, manipulators for nanoscale surgery, and drug reservoirs embedded in intelligent polymers are explored. All of these are virus-sized. The proliferation of these types of NBSs leads to a large number of applications, and combinations of these in the future will lead to microminiaturization, versatility, and functionality. Plant species present a great diversity of genetics, and wild ones constitute a large genetic reservoir, from which genes that are important from a practical point of view can be obtained. Plant genetic engineering research has a great theoretical significance, facilitating the knowledge of how genes of these organisms act, the effects of phytohormones on plant development, genes inactivation mechanisms, etc. By applying molecular biology techniques, useful information can be obtained on the genome of plants used for amelioration, the localization of genes of interest, the degree of relationship between different species, etc. As far as practical applications are concerned, a number of significant results have been obtained so far, some of which have already been applied, such as virus-resistant plants, plants resistant to the attack of pests, herbicide-resistant plants, plants of horticultural interest (ornamental plants with new phenotypes, plants producing softening resistant fruits), plants capable of synthesizing secondary metabolites in increased amounts, and plants producing “edible” antibodies, and enumeration could continue.
References
Affi M, Solliec C, Legentilhomme P, Comiti J, Legrand J, Jouanneau S, Thouand G (2016) Numerical modeling of the dynamic response of a bioluminescent bacterial biosensor. Anal Bioanal Chem 408(30):8761–8770
Agrawal R, Satlewal A, Chaudhary M, Verma A, Singh R, Verma AK, Kumar R, Singh KP (2012) Rapid detection of cadmium–resistant plant growth promotory rhizobacteria: a perspective of ELISA and QCM-based immunosensor. J Microbiol Biotechnol 22(6):849–855
Alvarez-Fernandez R (2010) Patented applications of gene silencing in plants: manipulation of traits and phytopathogen resistance. Recent Pat DNA Gene Seq 4(3):167–180
Armstrong W, Beckett PM (2011) Experimental and modelling data contradict the idea of respiratory down–regulation in plant tissues at an internal [O2] substantially above the critical oxygen pressure for cytochrome oxidase. New Phytol 190(2):431–441
Barroso MF, Freitas M, Oliveira MB, de-los-Santos-Álvarez N, Lobo-Castañón MJ, Delerue-Matos C (2015) 3D-nanostructured Au electrodes for the event–specific detection of MON810 transgenic maize. Talanta 134:158–164
da Silva CP, Franzoi AC, Fernandes SC, Dupont J, Vieira IC (2013) Development of biosensor for phenolic compounds containing PPO in β–cyclodextrin modified support and iridium nanoparticles. Enzym Microb Technol 52(4–5):296–301
Espinoza MA, Istamboulie G, Chira A, Noguer T, Stoytcheva M, Marty JL (2014) Detection of glycoalkaloids using disposable biosensors based on genetically modified enzymes. Anal Biochem 457:85–90
Fang X, Chen J, Dai L, Ma H, Zhang H, Yang J, Wang F, Yan C (2015) Proteomic dissection of plant responses to various pathogens. Proteomics 15(9):1525–1543
Frederickson Matika DE, Loake GJ (2014) Redox regulation in plant immune function. Antioxidant Redox Signal 21(9):1373–1388
Gaggeri R, Rossi D, Christodoulou MS, Passarella D, Leoni F, Azzolina O, Collina S (2012) Chiral flavanones from Amygdalus lycioides Spach: structural elucidation and identification of TNF alpha inhibitors by bioactivity–guided fractionation. Molecules 17(2):1665–1674
Genfa L, Jiang Z, Hong Z, Yimin Z, Liangxi W, Guo W, Ming H, Donglen J, Lizhao W (2005) The screening and isolation of an effective anti–endotoxin monomer from Radix Paeoniae Rubra using affinity biosensor technology. Int Immunopharmacol 5(6):1007–1017
Hamers D, van Voorst Vader L, Borst JW, Goedhart J (2014) Development of FRET biosensors for mammalian and plant systems. Protoplasma 251(2):333–347
He W, Yuan S, Zhong WH, Siddikee MA, Dai CC (2016) Application of genetically engineered microbial whole–cell biosensors for combined chemosensing. Appl Microbiol Biotechnol 100(3):1109–1119
Heyl A, Riefler M, Romanov GA, Schmülling T (2012) Properties, functions and evolution of cytokinin receptors. Eur J Cell Biol 91(4):246–256
Hines G, Modavi C, Jiang K, Packard A, Poolla K, Feldman L (2015) Tracking transience: a method for dynamic monitoring of biological events in Arabidopsis thaliana biosensors. Planta 242(5):1251–1261
Ibrahim MA, Stewart–Jones A, Pulkkinen J, Poppy GM, Holopainen JK (2008) The influence of different nutrient levels on insect–induced plant volatiles in Bt and conventional oilseed rape plants. Plant Biol (Stuttg) 10(1):97–107
Jacoby RP, Millar AH, Taylor NL (2015) Assessment of respiration in isolated plant mitochondria using Clark–type electrodes. Methods Mol Biol 1305:165–185
Jones AM, Grossmann G, Danielson JÅ, Sosso D, Chen LQ, Ho CH, Frommer WB (2013) In vivo biochemistry: applications for small molecule biosensors in plant biology. Curr Opin Plant Biol 16(3):389–395
Kazakova LI, Shabarchina LI, Anastasova S, Pavlov AM, Vadgama P, Skirtach AG, Sukhorukov GB (2013) Chemosensors and biosensors based on polyelectrolyte microcapsules containing fluorescent dyes and enzymes. Anal Bioanal Chem 405(5):1559–1568
Kersten B, Feilner T (2007) Generation of plant protein microarrays and investigation of antigen–antibody interactions. Methods Mol Biol 355:365–378
Knecht K, Seyffarth M, Desel C, Thurau T, Sherameti I, Lou B, Oelmüller R, Cai D (2010) Expression of BvGLP–1 encoding a germin–like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol Plant-Microbe Interact 23(4):446–457
Kozan JV, Silva RP, Serrano SH, Lima AW, Angnes L (2007) Biosensing hydrogen peroxide utilizing carbon paste electrodes containing peroxidases naturally immobilized on coconut (Cocos nucifera L.) fibers. Anal Chim Acta 591(2):200–207
Kurien BT, D'Souza A, Scofield RH (2010) Heat-solubilized curry spice curcumin inhibits antibody-antigen interaction in in vitro studies: a possible therapy to alleviate autoimmune disorders. Mol Nutr Food Res 54(8):1202–1209
Li X, Han B, Xu M, Han L, Zhao Y, Liu Z, Dong H, Zhang C (2014) Plant growth enhancement and associated physiological responses are coregulated by ethylene and gibberellin in response to harpin protein Hpa1. Planta 239(4):831–846
Liu LH, Zhou XH, Shi HC (2015) Portable optical aptasensor for rapid detection of mycotoxin with a reversible ligand–grafted biosensing surface. Biosens Bioelectron 72:300–305
Liu X, Chen M, Onstad D, Roush R, Shelton AM (2011) Effect of Bt broccoli and resistant genotype of Plutella xylostella (Lepidoptera: Plutellidae) on development and host acceptance of the parasitoid Diadegma insulare (Hymenoptera: Ichneumonidae). Transgenic Res 20(4):887–897
Lukács A, Garab G, Papp E (2006) Measurement of the optical parameters of purple membrane and plant light–harvesting complex films with optical waveguide lightmode spectroscopy. Biosens Bioelectron 21(8):1606–1612
McLamore ES, Jaroch D, Chatni MR, Porterfield DM (2010) Self–referencing optrodes for measuring spatially resolved, real–time metabolic oxygen flux in plant systems. Planta 232(5):1087–1099
Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R (2007) Redox–sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52(5):973–986
Michelini E, Roda A (2012) Staying alive: new perspectives on cell immobilization for biosensing purposes. Anal Bioanal Chem 402(5):1785–1797
Miranda OR, Li X, Garcia–Gonzalez L, Zhu ZJ, Yan B, Bunz UH, Rotello VM (2011) Colorimetric bacteria sensing using a supramolecular enzyme–nanoparticle biosensor. J Am Chem Soc 133(25):9650–9653
Muñoz FJ, Rumbero A, Sinisterra JV, Santos JI, André S, Gabius HJ, Jiménez-Barbero J, Hernáiz MJ (2008) Versatile strategy for the synthesis of biotin–labelled glycans, their immobilization to establish a bioactive surface and interaction studies with a lectin on a biochip. Glycoconj J 25(7):633–646
Nelsen B, Kadesch T, Sen R (1990) Complex regulation of the immunoglobulin mu heavy-chain gene enhancer: microB, a new determinant of enhancer function. Mol Cell Biol 10(6):3145–3154
Nikolelis DP, Ntanos N, Nikoleli GP, Tampouris K (2008) Development of an electrochemical biosensor for the rapid detection of naphthalene acetic acid in fruits by using air stable lipid films with incorporated auxin–binding protein 1 receptor. Protein Pept Lett 15(8):789–794
Panchal MB, Upadhyay SH (2014) Boron nitride nanotube-based biosensing of various bacterium/viruses: continuum modelling–based simulation approach. IET Nanobiotechnol 8(3):143–148
Potocký M, Pleskot R, Pejchar P, Vitale N, Kost B, Zárský V (2014) Live–cell imaging of phosphatidic acid dynamics in pollen tubes visualized by Spo20p–derived biosensor. New Phytol 203(2):483–494
Prasad R, Bhattacharyya A, Nguyen QD (2017) Nanotechnology in sustainable agriculture: recent developments, challenges, and perspectives. Front Microbiol 8:1014. https://doi.org/10.3389/fmicb.2017.01014
Prasad R, Kumar V, Prasad KS (2014) Nanotechnology in sustainable agriculture: present concerns and future aspects. Afr J Biotechnol 13(6):705–713
Ramirez RA, Spears LR (2014) Stem nematode counteracts plant resistance of aphids in alfalfa, Medicago sativa. J Chem Ecol 40(10):1099–1109
Raymond B, Sayyed AH, Wright DJ (2007) Host plant and population determine the fitness costs of resistance to Bacillus thuringiensis. Biol Lett 3(1):82–85
Richard MM, Pflieger S, Sévignac M, Thareau V, Blanchet S, Li Y, Jackson SA, Geffroy V (2014) Fine mapping of Co–x, an anthracnose resistance gene to a highly virulent strain of Colletotrichum lindemuthianum in common bean. Theor Appl Genet 127(7):1653–1666
Rodríguez–Sevilla E, Ramírez–Silva MT, Romero–Romo M, Ibarra–Escutia P, Palomar–Pardavé M (2014) Electrochemical quantification of the antioxidant capacity of medicinal plants using biosensors. Sensors (Basel) 14(8):14423–14439
Sakamoto S, Putalun W, Vimolmangkang S, Phoolcharoen W, Shoyama Y, Tanaka H, Morimoto S (2018) Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J Nat Med 72(1):32–42
Stein NE, Keesman KJ, Hamelers HV, van Straten G (2011) Kinetic models for detection of toxicity in a microbial fuel cell based biosensor. Biosens Bioelectron 26(7):3115–3120
Takakusagi Y, Manita D, Kusayanagi T, Izaguirre–Carbonell J, Takakusagi K, Kuramochi K, Iwabata K, Kanai Y, Sakaguchi K, Sugawara F (2013) Mapping a disordered portion of the Brz2001–binding site on a plant monooxygenase, DWARF4, using a quartz–crystal microbalance biosensor–based T7 phage display. Assay Drug Dev Technol 11(3):206–215
Vigneux F, Zumbihl R, Jubelin G, Ribeiro C, Poncet J, Baghdiguian S, Givaudan A, Brehélin M (2007) The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J Biol Chem 282(13):9571–9580
Wan Salim WW, Zeitchek MA, Hermann AC, Ricco AJ, Tan M, Selch F, Fleming E, Bebout BM, Bader MM, Ul Haque A, Porterfield DM (2013) Multi-analyte biochip (MAB) based on all-solid-state ion–selective electrodes (ASSISE) for physiological research. J Vis Exp (74)
Werner S, Polle A, Brinkmann N (2016) Belowground communication: impacts of volatile organic compounds (VOCs) from soil fungi on other soil–inhabiting organisms. Appl Microbiol Biotechnol 100(20):8651–8665
Wolfbeis OS (2015) Luminescent sensing and imaging of oxygen: fierce competition to the Clark electrode. BioEssays 37(8):921–928
Wu H, Liu J, Zhang J, Li C, Fan J, Xu X (2014) Comparative quantification of oxygen release by wetland plants: electrode technique and oxygen consumption model. Environ Sci Pollut Res Int 21(2):1071–1078
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Butnariu, M., Butu, A. (2019). Plant Nanobionics: Application of Nanobiosensors in Plant Biology. In: Prasad, R. (eds) Plant Nanobionics. Nanotechnology in the Life Sciences. Springer, Cham. https://doi.org/10.1007/978-3-030-16379-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-16379-2_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-16378-5
Online ISBN: 978-3-030-16379-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)