Abstract
Single-particle cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image processing have gained importance in the last few years to obtain atomic structures of drug targets. Obtaining atomic-resolution 3D structure better than ~2.5 Å is a standard approach in pharma companies to design and optimize therapeutic compounds against drug targets like proteins. Protein crystallography is the main technique in solving the structures of drug targets at atomic resolution. However, this technique requires protein crystals which in turn is a major bottleneck. It was not possible to obtain the structure of proteins better than 2.5 Å resolution by any other methods apart from protein crystallography until 2015. Recent advances in single-particle cryo-EM and 3D image processing have led to a resolution revolution in the field of structural biology that has led to high-resolution protein structures, thus breaking the cryo-EM resolution barriers to facilitate drug discovery. There are 24 structures solved by single-particle cryo-EM with resolution 2.5 Å or better in the EMDataBank (EMDB) till date. Among these, five cryo-EM 3D reconstructions of proteins in the EMDB have their associated coordinates deposited in Protein Data Bank (PDB), with bound inhibitor/ligand . Thus, for the first time, single-particle cryo-EM was included in the structure-based drug design (SBDD) pipeline for solving protein structures independently or where crystallography has failed to crystallize the protein. Further, this technique can be complementary and supplementary to protein crystallography field in solving 3D structures. Thus, single-particle cryo-EM can become a standard approach in pharmaceutical industry in the design, validation, and optimization of therapeutic compounds targeting therapeutically important protein molecules during preclinical drug discovery research. The present chapter will describe briefly the history and the principles of single-particle cryo-EM and 3D image processing to obtain atomic-resolution structure of proteins and their complex with their drug targets/ligands.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The importance of structural biology in understanding the principles of molecular function of proteins, the workforce of cellular world, underpins its use in health science and pharma industries. Classically, protein crystallography was ruling the world of structure-based drug design (SBDD). This was mainly due to the capability of protein crystallography to solve high (better than 1.8 Å), atomic (better than 1.2 Å), and ultra-high (better than 0.95 Å)-resolution 3D structures, which give information of protein drug molecular interaction at various levels. Particularly, the positions of hydrogen atoms were located in many atomic and ultra-high-resolution protein structures. There were no other methods that could rival the versatility of obtaining 3D atomic-level macromolecular structures with which crystallography could achieve. Of the 131,108 protein structures in PDB (as on June 15, 2018), 90% of structures among them were solved by X-ray crystallography technique and 8% by NMR technique. The remaining 2% of structures by large were solved by electron microscopy, electron crystallography, hybrid, and other methods, which include neutron diffraction, solution scattering, fiber diffraction. Clearly, the PDB data suggests that the protein crystallography technique dominates till date. However, the protein crystallography method comes with a proviso. That is, we need diffractable protein crystals of reasonable 10–100s of micron size, in order to obtain a high-resolution X-ray crystallography protein structure. Also, as the unit cell parameter of the protein crystals increase, the resolution of diffraction data drops as the cube of unit cell parameter [1]. Moreover, many proteins, in particular membrane proteins and fibrous proteins, are recalcitrant to crystallization. An analysis of deposited protein structures in PDB by Kozma and co-workers in 2017 [2] showed that the majority of the solved structures (97.6%) are globular proteins and only ~2.4% of them are membrane protein structures. This is primarily because obtaining good diffraction quality 3D crystals for membrane proteins is challenging. As a result, single-particle cryo-EM has gained popularity nowadays for solving membrane protein structures as well along with globular proteins. Also, in cases where single-particle cryo-EM cannot give high-resolution maps, protein crystallography and cryo-EM can be used as hybrid method to visualize macromolecular assemblies at pseudo-atomic resolution as described in Natesh [3] and references cited therein.
SBDD is among one of the most important stages for drug discovery in industrial drug discovery pipelines [4]. It requires the best possible resolution protein structures, preferably better than 2.5 Å resolution. Until 2015, single-particle cryo-EM could not achieve the resolution comparable to resolution of structures in protein crystallography [5, 6]. Recently, Danev and co-workers have solved a structure of Mus musculus apo ferritin at 1.62 Å (EMD-9599). Others have solved the structures of proteins with bound ligands at resolution 2.5 Å or better [7,8,9,10], presented in Table 1. The foundation for this was laid 36 years ago in December 1981 when Jacques Dubochet (along with AW Mc Dowall) published the paper on vitrification (amorphous ice) of pure water for electron microscopy [11]. Jacques was excited about the prospects of making electron microscopy water friendly. Five years after that, they got the first cryo-EM virus structure at 35 Å resolution [12]. However, before that the first EM structure came from Henderson and Unwin [13] of purple membrane protein by electron crystallography, but however not using cryo, and hence, the resolution was bit low at 7 Å. This encouraged Joachim Frank to develop image processing algorithms for solving protein structures by building 3D reconstruction from fussy cryo-EM projection images of proteins [14,15,16]. These developments led to the first cryo-EM atomic model of the protein bacteriorhodopsin 15 years later in the year 1990 [17]. In recent years, other developments like field emission gun electron source, direct electron detectors, and movie-based cryo-EM imaging methods have led to an avalanche of high-resolution single-particle cryo-EM protein structures [5, 6, 18]. Thus, the full potential of cryo-EM in obtaining high-resolution structure of proteins was realized in 2015, which led to the Noble Prize in Chemistry in the year 2017 for “developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution.” The predictions made by Henderson in 1995 [19] that single-particle cryo-EM can be used for atomic-resolution structure determination of protein and protein complexes has become a reality today. Thus, single-particle cryo-EM technique can be used as a pipeline for obtaining atomic structures of druggable targets in preclinical SBDD.
2 The Single-Particle Cryo-EM at High Resolution
The single-particle cryo-EM method for high-resolution structure determination of proteins and protein complexes involves four major steps, viz. (i) the sample preparation, (ii) specimen preparation, (iii) data collection, and (iv) image processing and 3D reconstruction (i.e., structure determination, which includes model building and refinement of the protein/ligand coordinates in the EM map). Sample preparation involves protein purification either from the source or expressed recombinantly in a heterologous host system. The amount of sample required for cryo-EM is very less (~1 µM) in comparison with protein crystallography or NMR spectroscopy techniques, where typically ~200 µM sample is required.
For single-particle electron microscopy (EM), there are two main ways of specimen preparation: (a) negative stain specimen preparation and (b) solution-state “vitrification” for cryo-EM. The former is used for quick characterization of macromolecules and their complexes. However, this type of specimen preparation involves inherent drawbacks (e.g., artifacts and visualizing stain rather than actual protein), which limits the resolution of EM reconstruction map from 30 to 20 Å at its best. Single-particle cryo-EM, the focus of this chapter, on the other hand is synonymous to solution-state structure, and the specimen preparation does not induce artifacts over the protein sample being studied. The vitrified specimen preserves the resolution of the protein structure that is being studied.
Single-particle cryo-EM technique has the capability to solve protein structures to better than 4 Å resolution nowadays. It is to be noted that, there is a consensus in the EM community that better then 4 Å depicts high-resolution structures, while, in the X-ray crystallography community, high resolution corresponds to better than 1.8 Å resolution, as described in the beginning of this chapter. Prior to the resolution revolution in the year 2015, most of the cryo-EM structures with resolution 4 Å or better were virus structures [20,21,22]. This was possible due to their large size and high symmetry (e.g., icosahedron symmetry). Most of these data were collected on photographic film (KODAK SO-163 FILM). However, the asymmetric particles (i.e., particles without higher-order symmetry) were limited to sub-nanometer (around 6–10 Å) resolution. Only 1/10th of the total number of structures in EMDB were with resolution 4 Å or better before the resolution revolution. This has significantly increased to 1/6th of the total number of single-particle cryo-EM structures in EMDB as on July 29, 2018, clearly indicating that, currently, there are more structures solved with resolution better than 4 Å in the database. These were possible due to the advancement in the hardware and software and the way the projection images are captured and processed during cryo-EM data collection and processing. Main steps involved in single-particle cryo-EM for obtaining high-resolution protein structure are presented in three subsections. First, we will begin with the details of the specimen preparation in Sect. 2.1, followed by data collection in Sect. 2.2, and finally image processing and 3D reconstruction in Sect. 2.3, respectively.
2.1 Specimen Preparation for Single-Particle Cryo-EM
The cryo-EM specimen preparation is the most challenging and a crucial step for high-resolution data collection. Most of the time spent on single-particle cryo-EM pipeline is in preparing the best protein specimen (which involves optimizing both the biochemistry and vitrification of the sample) for high-resolution cryo-EM data collection. Hence, it is worth to spend some time to get the best specimen out from the purified sample, which will save time and money later. The first step in specimen preparation is the purified sample (e.g., protein or protein complexes) typically 3–3.5 µL is applied on a pre-glow discharged holey carbon grid (in some special cases, continuous carbon grids laid over the holey grid are used). For the best specimen preparation, the quality of the protein sample, pre-treatment of holey grids, and the choice of the type of grid are important. After applying the sample on the holey carbon grid, the excess protein is blotted using a filter paper (usually Whatman filter paper 1) to leave a very thin layer of sample and immediately the grid is plunged into a pre-prepared liquid ethane well, surrounded and maintained at cryogenic temperature by a bath (surrounding the ethane well) with liquid nitrogen as shown in Fig. 1. Jacques Dubochet and co-workers standardized the vitrification process in the late 1970s and published the work in the year 1981 [11]. They showed that sample in buffer/water must be cooled in less than a millisecond to avoid the ice crystal formation and to get amorphous ice (i.e., vitrified). They also showed that if the temperature of specimen is kept sufficiently low below −160 °C, the vitrified state could be maintained for long time [11, 23]. This seminal discovery enabled proteins to be visualized in its native state under the vacuum of transmission electron microscope (TEM). For this discovery, Dubochet received one-third of the Noble Prize in Chemistry in the year 2017. The solution-state protein sample is frozen in time and space, maintaining the integrity of the protein’s structural state in the vitrified water. The vitrification can be carried out with a homemade manual plunger or using a commercially available vitrification robot. A perfect vitrified specimen is one in which the thickness of the ice over the holes of the grid is such that there is one single layer of particles distributed, the particles are uniformly distributed (with distance between each particles at least 1.5 times the particle size), and the particles adopt as many different orientations as possible. The vitrified specimen grid is then placed in a cryo grid storage box that is preserved in liquid nitrogen storage Dewar, until the data collection is carried on a high-resolution cryo-TEM. An extensive description of the specimen preparation is given in Passmore and Russo [24].

Figure reproduced from Natesh, 2014 [3], by permission of publisher—Indian Academy of Science, Bengaluru
Vitrification of cryo-EM specimens. A cryo-EM grid with a thin film of solution (<2000 Å of thickness) is plunged into liquid ethane for vitrification. The frozen specimen is transferred into liquid nitrogen before it is imaged at liquid nitrogen temperatures on a TEM. To the right top is a schematic edge-on view of a part of frozen water layer, with macromolecular complexes trapped in different orientations. Bottom right, part of a cryo-EM image showing weak and noisy views of the complexes.
2.2 Data Collection
Data collection is carried out on a cryo-TEM equipped with a 200 or 300 kV field emission gun (FEG) necessary to obtain a high-resolution single-particle data. The stored grids are transferred from the cryo grid storage box to a single tilt cryo-transfer holder pre-cooled on a cryo-workstation (Fig. 2b). In this case, only one grid can be inserted into the TEM by manually loading the holder into the cryo-TEM (Fig. 2a) and analyzed before the holder is taken out of the microscope at the end of data collection. Alternatively, each one of the stored grids can be transferred one by one to a cartridge, which is then placed on multiple grid holder cassette (which holds up to 12 grids). This cassette is then placed into the capsule, which is loaded into the cryo-TEM (Fig. 2c) through an autoloader robot that is built in the microscope. Thermo Fisher Scientific Talos Arctica/Glacios, Thermo Fisher Scientific Krios, and JEOL Cryo ARM 200/300 are microscopes with such autoloader capabilities. The robotic grid loader then can load one by one to the stage using inbuilt robot, which can load or unload the grid on the stage controlled by software. In case of high-end TEM analysis, grid atlas can be created to choose the square of right thickness from all the loaded grids. It is very important to keep the grid always under liquid nitrogen in order to avoid any ice crystal formation and contamination on the grid. Hence, all the processes described in Fig. 2, which involve handling of frozen specimen grid, are carried out under liquid nitrogen. Ice crystals destroy the view of particles by dark contrast, and hence, it is critical to avoid any exposure of plunge-frozen grid to the air.
a 200 kV transmission electron microscope equipped with field emission gun (FEG). b Gatan CT3500 single tilt liquid nitrogen cryo-transfer holder docked onto cryo-workstation. After inserting the specimen grid onto the cryo-holder (not in scale to microscope), it is carefully transferred to the microscope as shown by the arrow mark. c A maximum of 12 grids can be loaded via cassette, housed in a capsule as described in the text. Each grid can be imaged one by one using an autoloader robot housed in a 200- or 300-kV cryo-TEM. (Fig. 2a, b was reproduced from Natesh [3], by permission of publisher—Indian Academy of Science, Bengaluru)
Once the grid is on the stage of the TEM, the data is collected on a highly sensitive direct detection device (DDD) also called as direct electron detector (DED) under low electron dose (typically <15 e−/Å2). Low electron dose is necessary since high dose (>15–20 e−/Å2) will cause radiation damage. However, high dose ~1000 e−/Å2 is required for atomic-resolution reconstruction [25]. This problem can be overcome by averaging similar looking particles as described in image processing Sect. 2.3 below. DDD is more sensitive (technically, this feature is called improved detective quantum efficiency (DQE)) and can detect lower doses more effectively with low noise as compared to the conventional photographic film or the CCD (charged coupled device) detectors. Data collection at the focus gives the best resolution, but however the phase contrast is lost in the image (i.e., you cannot clearly visualize the particles). In order to visualize the particles, the images are captured at a defocus that restores the phase contrast in the image, which enables us to visualize particles. Hence, data is collected at a range of defocus between ~4 µm (lower resolution) and ~1 µm (higher resolution). Modern-day advancements in hardware have led to the use of phase plates and energy filters that can restore contrast in the images collected closer to focus. Thus, preserving high-resolution information in the images and at the same time preserving the image phase/amplitude contract as a result alleviate the need for contrast transfer function (CTF) modulation correction at image processing stage.
For high-resolution structure determination, the data is collected on DED as movie frames, which is actually a dose fractionated image stack. The movie frames collected can be corrected for loss of resolution due to stage drifts, charging, and beam-induced motion. The individual movie frames or subset of movie frames in batches are then aligned with respect to each other in order to restore the high-resolution information [26]. Relatively, high exposures up to 20 e−/Å2 can be used for movie mode while DEDs can also be used in electron counting mode where dose rate must be kept below 10 e−/pixel/s [26, 27]. Movie corrections are applied immediately on the micrographs after the data collection using programs like MotionCor2 [28], optical flow algorithm as implemented in Xmipp [29, 30], Unblur/Summovie [31, 32]. In addition, improved stability of specimen can be provided by the use of grids with graphene and gold support [25, 33, 34]. Hence, in the last six years there has been many breakthroughs in detector, imaging, and image processing technology that has led to high-resolution data collection for even smaller proteins like hemoglobin with mass 64 kDa using Volta phase plate (VPP) [18], thus leading to resolution revolution with structures determination to better than 2.5 Å. Another aspect of data collection is the automation. Not all proteins give homogenous samples for atomic-resolution reconstruction. The fact that proteins are dynamic leads to heterogeneity and underlies the need for large amount of data collection (in a hope to group particles into homogenous groups), which is tedious to be done manually. In recent years, many software packages have been developed to interface with the advanced electron microscopes for automatic data acquisition. Some examples of such software that can be used for fully automated data collection on a well-calibrated cryo-TEM are Leginon [35], SerialEM [36], UCSFImage4 [37], FEI-EPU, JEOL-JADAS [38], GATAN-Latitude S. Most of the software is used for automated data collection for both single-particle cryo-EM and electron tomography (ET) work. Some programs like Appion [39] extend the automated data collection through a pipeline from automated data collection all the way through automated particle picking to image processing (CTF estimation, classification, and 3D reconstruction).
2.3 Image Processing and Three-Dimensional Reconstruction
Cryo-EM is different from X-ray crystallography because it uses “images” as primary data, rather than the diffraction patterns. Translated into Fourier lingo, the availability of images means that the “phase problem” known in X-ray crystallography (described in Sects. 2.3 and 2.4 of Natesh [3]) does not exist in EM. The electron microscope, in Hoppe’s words, is a “phase-measuring diffractometer” [40]. Hence, extreme care has to be taken in image processing. Image processing involves preprocessing the collected data, particle picking, centering the particles in their selected boxes, 2D classification and determining their relative orientations and/or 3D classification and 3D reconstruction. An example of image processing and 3D reconstruction is shown in Figs. 3 and 4. The preprocessing step involves CTF correction and image normalization [41]. As mentioned in the data collection section, the data is collected at various defocus positions. As one gradually increases the defocus (i.e., under focus), the contrast of the image proportionally improves. Improvement in contrast comes at a cost, a loss in the higher spatial frequencies (i.e., high-resolution information is lost) in the image, and in addition, it introduces CTF modulation in the spatial frequencies of the image. Hence, the first step in image processing is to calculate the lens defocus and astigmatism, which is needed to correct the measured data for the CTF of the microscope [42, 43]. Software CTFFIND, ACE2, Gctf, or e2ctf.py [44,45,46,47] can be used to estimate the CTF that is used for CTF corrections.
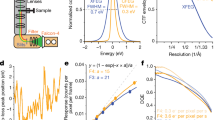
Image processing and 3D reconstruction of GroEL and non-native protein RuBisCO complex [74]. a Raw micrograph (this image is not motion corrected, but at this stage if dose fractionated image stacks are collected on a DED, they are motion corrected) and b 30-Å-filtered brick view reference from empty GroEL cryo-EM map. (c) Particles from cryo-EM images like in micrograph (a) are extracted into boxes, CTF corrected, filtered, normalized, and aligned to reference to bring them to the same center. d Orientation separation by class averages of images using MSA shows significant improvement in signal-to-noise ratio. e Eigen image information (circled) was used to classify images into homogenous classes. f MSA classification into three homogenous groups; 3D reconstruction of three classes using projection matching is shown in Fig. 4

Figure produced by the author in https://doi.org/10.1016/j.ijbiomac.2018.06.120 [74] and reproduced here under Creative Commons Attribution License (CC BY)
Asymmetric (C1) 3D reconstructions of the three classes (structures). Each class (class 1 (a); class 2 (b); and class 3 (c)) is shown as a top view (top ring only), a side view, a central section through the side view, and a bottom view (bottom ring only). The fitted GroEL crystal structure is shown in green. The additional density in the upper rings of (a) and (b) is attributable to bound non-native RuBisCO substrate. All maps were contoured at the 1σ level without filtering. Figure generated with Chimera [85].
After CTF correction, the images are normalized to set the mean density of the particles to zero and same standard deviation [41]. The particles are then manually or auto-picked into boxes of 1.5–2.5x, the size of the largest axis of the particle using suitable software. A guide for choosing the right box size is given at the online documentation http://blake.bcm.edu/emanwiki/EMAN2/BoxSize. Number of softwares are available for manual and automatic picking of particles and subsequent image processing. Examples of such programs are FindEM [48] (only for automated particle picking), EMAN (e2boxer.py) [49, 50], IMAGIC [51], Ximdisp [52] (only for interactive display, analyses, and particle picking; now a part of CCP-EM package [53, 54]), Xmipp [30], RELION-autopick [55], cryoSPARC [56], APPLE Picker [57] (completely automatic particle picking, a part of ASPIRE Suite [58]), gEMpicker [59] (only for template-based particle picking), SIGNATURE [60] (only for particle picking and data analysis), etc. Most of the auto-picking software employ initial manual picking routine (except APPLE picker), where a couple of thousands of particles are manually picked from a subset of available micrographs and use the best class averages generated from them (having as many different representative orientations) as templates to auto-pick particles from rest of the micrographs. This is the preferred method. Alternatively, the auto-picking programs can use low-pass-filtered EM maps as templates for particle picking (less preferred, but useful in protein drug complex where you have the apo-protein structure already). Using maps from PDB (Protein Data Bank) coordinates, as reference model is not preferred at this stage in order to avoid “Einstein-from-noise” effect [61], i.e., to avoid any 2D model bias. CTF corrections can also be performed on picked particle images as compared to whole micrographs in some software, e.g., EMAN [44].
After particle picking, the next stage is to get the 3D reconstruction of the biological macromolecules using the different but identifiable 2D projections of particles. The first 3D reconstruction from a 2D projection was carried out on negative stained tail of bacteriophage T4 by De Rosier and Klug [62]. However, the 2D projections of particle images cutout from the motion-corrected micrographs have still low signal-to-noise ratio (SNR) due to low electron dose data collection as described in data collection section. Hence, in order to improve the SNR of the particles, many identical looking particle images are aligned and summed (clustering) thus effectively increasing the SNR and dose without increasing the damage [62]. There are three main advantages of reference-free (unsupervised) 2D classification: (i) to select few 2D classes from which we can make starting 3D map, which can be projected as references for refinement. (ii) We can identify the fraction of bad classes (which may contain artifacts, invalid particles, or simply empty), and thus, those images with anomalies can be deleted from the data set in the beginning itself. (iii) It also helps in identifying the conformational and compositional variability in the data set [50]. Two-dimensional (2D) and 3D classifications are carried out by using various statistical analysis software suite IMAGIC [51], Spider [63], EMAN [44], RELION-3 [64], FREALIGN [65], Appion [39], cryoSPARC [56], ASPIRE Suite [58], Xmipp [30], SPHIRE (sphire.mpg.de), etc., or a combination of more than one of these suites. Several of these software packages are integrated into one processing framework , for example, as in Scipion [66]. An exhaustive list of EM software programs is available at EMDataBank (EMDB, http://www.emdatabank.org/emsoftware.html).
Spider [63, 67] and IMAGIC [51] were among the first programs to be developed for single-particle reconstruction in the year 1996 followed by FREALIGN [65] in the year 1998 and other program suites followed. The clustering of similar particle images was first introduced by van Heel and Frank [68] in the year 1981 using multivariate statistical analysis. Clustering in the currently available programs uses one of the following methods: multivariate statistical analysis (MSA)/principle component analysis (PCA), hierarchical clustering, k-means clustering, and the maximum-likelihood methods [41] or by recently proposed empirical Bayesian approach [69]. Currently, the EMAN2 [49] and RELION-3 [64] are among the popular program that do reference-free 2D class-averaging (references are generated from within the data set) and 3D reconstruction. EMAN2 uses iterative MSA-based reference-free 2D classification. The latest one, the RELION, uses empirical Bayesian likelihood approach for 2D classification [55].
Next step is to get the 3D reconstruction from selected good class averages. High-resolution 3D reconstructions require an initial 3D model that can be iteratively refined to obtain the best possible resolution for the data set. The first starting 3D model is obtained using experimental methods or by finding the relative orientations of 2D projection averages (and hence the particles) by computational methods . Assigning orientations by programs involves finding the location and Euler angles of the particles in the boxed region. The earliest one among them was the popular angular reconstitution method [70] by Marin van Heel, which uses real-space implementation of “common lines” principle to get relative orientations of the class averages as implemented in the program IMAGIC [51]. Thus, the Euler angles assigned 2D class averages can be used to get the starting 3D model. This method does not require reference for assigning relative orientation, while another program Spider by Joachim Frank and co-workers uses projection matching and cross-correlation approach [63, 71]. This method requires a starting 3D model which is generated from ab initio random conical tilt method [72] from EM images taken at a pair of know angles. Most of the present-day programs generate the starting 3D model by using statistical approach and comparison with back-projections to assign the Euler angles to a subset of manually selected good class averages. For example, EMAN2 uses a Monte Carlo method, RELION uses Bayesian methods, and VIPER [73] a module in SPHIRE suite (http://sphire.mpg.de/) uses a stochastic hill-climbing algorithm. Iterative rounds of projection matching with the references generated from starting 3D model (called as 3D projection matching procedure) followed by subsequent 3D reconstruction (using various algorithms) are used until the resolution of the reconstruction during subsequent refinement cycles does not further improve. This will lead to the final 3D reconstruction with the best possible resolution.
Figure 4 shows an asymmetric (C1 symmetry) 3D reconstruction carried out using IMAGIC and Spider. The non-native RuBisCO bound to GroEL is shown [74]. Figure 5 is another example of 1.9 Å high-resolution cryo-EM reconstruction with inhibitor phenylethyl β-D-thiogalactopyranoside (PETG) bound to β-galactosidase enzyme [75]. The quality of the final 3D reconstruction not only depends on the quality of the projection images and implementation of the clever algorithms, but also on the angular distribution of the particles. Hence, in order to get the best resolution reconstruction, it is necessary for the particle (and thus its projections) to be distributed well in the Euler sphere [41]. By re-projecting the 3D reconstruction at the Euler angles of the class averages, we can assess the reliability of the 3D reconstruction. For a consistent reliable reconstruction, the re-projected image and the actual class average must match.
Fourier shell correlation (FSC) curve for class 1, class 2, and class 3 asymmetric reconstruction and class 3 (C7 symmetry reconstruction) shown in Fig. 4. Vertical dashed lines show the spatial frequency for 0.143 “gold-standard” FSC which estimates classes 1, 2, and 3 resolution to be ~9.0 Å and class 3 (C7 symmetry) as ~7.6 Å
3 Resolution, Model Building, and Validation
3.1 Resolution
Resolution estimation of the EM maps is still subjective, with differences among various groups still not settled [76]. Resolution of 3D EM map is calculated from a plot of Fourier shell correlation (FSC) [77] as a function of spatial frequency (the resolution estimation of 3D reconstructions in Fig. 4 is shown in Fig. 5). FSC is the cross-correlation (CC) calculated between two 3D reconstruction maps, where each map is calculated from half the data images. The resolution that is reported in publication essentially as a single number is the value of maximum spatial frequency up to which the EM map is reliable. The identification of resolution is subjective as it is arbitrary what one considers as reliable. The procedure for resolution assessment is described in detail by Penczek [76]. There are several suggestions for identifying the cutoff: (i) the 3-sigma criteria where the spectral SNR (SSNR) = 0 in which case FSC = 0; (ii) point at which power of signal is equal to the power of noise, i.e., SSNR = 1 or FSC = 0.33; (iii) the classic midpoint of FSC curve, i.e., FSC = 0.5 [78] where SSNR = 2, which means signal dominates noise; and finally (iv) point where FSC = 0.143, derived by Rosenthal and Henderson [79] in comparison with X-ray crystallography. Hence, which cutoff is chosen is a matter of present-day debate. Recently, in order to reduce further any possible reference bias, “gold-standard FSC” was suggested with FSC calculated between two completely independent refinements and 3D reconstruction [80].
There are other computational ways to improve the resolution nominally without improving the image alignment, e.g., masking/threshold flattening. In any case, the resolution estimations have their own limitations and hence reported EM resolution should be treated as only broad guideline rather than a definitive number and cannot be used as validation. Nonetheless, it is an important parameter to be reported with each EM map deposition at the EMDB . Resolution anisotropy is common in cryo-EM structures, and it is a common practice to document it as color ramping from low to high resolution on the cryo-EM 3D reconstruction map using programs ResMap [81] and blocres [82]. The results can be visualized independently or with chimera (e.g., blocres with Local FSC plug-in for chimera).
With the booming medium- and high-resolution cryo-EM 3D structures, it is necessary to have consistency between crystallography and cryo-EM terms currently used for defining what is an atomic- or high-resolution structure. While it is very common to use the term “atomic resolution” for cryo-EM resolutions better than 3.5 Å, the crystallography definition of the term “atomic resolution” means the resolution is 1.2 Å or better [1] and ultra-high resolution means 0.95 Å or better ([83] and references cited therein). Similarly, 1.8 Å or better is called high resolution [84], 3.0 Å or better up to 1.9 Å is treated as medium resolution while low resolution is between 4 and 3.1 Å. Resolution below 4 Å is considered as poor resolution in protein crystallography. While the method of estimation of resolution is quite different between crystallography and cryo-EM techniques, the conventions for using the terms should be consistent, irrespective of the method. Hence, the author would like to suggest that it is necessary for the cryo-EM field to maintain consistency in the future, while using the terms ultra-high, atomic, high, medium, and low resolution.
3.2 Model Building
If the resolution of the 3D reconstruction (i.e., the electron potential map) is sufficiently high, e.g., better than 3 or 4 Å, it is often possible to build ab initio atomic model and do refinement with the EM map using known chemical constraints/restraints. If X-ray crystallography coordinates of the segment or its homologues are available, one can rigid body fit the segment coordinates into the cryo-EM map using programs like UCSF Chimera [85]. Where the resolution of the 3D reconstruction map is limited to worse than 4 Å, combining crystallography and cryo-EM as a hybrid method is a powerful tool to obtain a pseudo-atomic model(s). Iterative rounds of model building using programs like Coot [86], O [87] and refinement using programs like Coot, refmac [88], or PHENIX real-space-refinement [89] are carried out. De novo backbone tracing and model building can be carried out using programs like Pathwalking and Gorgon [90]; it can also build macromolecular assemblies at non-atomic resolution [90]. When the cryo-EM map shows variation by domain movements or flexibility to the available protein coordinates, programs like FlexEM [91, 92] and MDFF [93] with its graphical user interface VMD [94] can be used to flexibly fit the coordinates in the EM map. The fitted model and EM map can be visualized in programs like PyMOL [95]/Chimera [85] to generate publication quality figures.
3.3 Validation
Validation in cryo-EM reconstruction is important to avoid errors in particle alignment, reference bias, over-fitting of atomic coordinates, and over-estimation of resolution. Validation tools for cryo-EM similar to the free R value (Rfree) in X-ray crystallography [96] have been introduced in 2003 by Joachim and co-workers [97] and recently by Chen and co-workers [98]. There are some general guidelines [80, 99] suggested, and they are actively evolving. As described in the resolution paragraph of this section, the reporting of resolution with one number cannot be used as validation; however, it is an important parameter to be reported during EMDB deposition. Further, FSC may fail when the particles are significantly misaligned. So one has to estimate the resolution properly [76] and use the reported single number resolution with caution. It is suggested that gold-standard FSC provides a realistic estimate of the true signal [100], and this will lead ultimately to a better map. In recent days, reporting local resolution has also formed a common practice in publication and thesis [81, 82]. Also, the local resolution will be helpful in avoiding over-interpretation of poor regions in the cryo-EM map. If the 3D map is of sufficient resolution (better than 4 Å), it can resolve the secondary structural features. A good validation would be especially if you can see a right-handed alpha helix or even the side chain residues, especially the bulky residues like tryptophan, phenylalanine, or tyrosine in the high-resolution cryo-EM map. Even the comparison of the new EM structure with the available EM structure will be one way of validating the newly reconstructed 3D EM map [101]. Further, programs TEMPy [102] and refmac [88] can be used to assess the validity of the fitted coordinates to the EM map. In Coot [86] program, one can use the Ramachandran plot (Validate →Ramachandran plot) and geometrical quality (Validate →Geometry analysis) to validate the quality of the refined model. One more way to validate is to compare the 3D reconstruction results from different techniques, e.g., projection matching and the angular reconstitution. For low-resolution maps (worse than 4 or 10 Å), measure of confidence can be provided by a priori random conical tilt experiments [103].
4 Heterogeneity
Though cryo-EM can handle heterogeneous particles, we need homogenous particles, which are equally dispersed in the vitrified ice in order to achieve atomic resolution. Ideally speaking, all data sets are heterogeneous! The question is how much one is willing to tolerate [104]. Further, during cryo-EM specimen preparation, non-physiological structural heterogeneity is often introduced [105]. While structural heterogeneity is a problem to obtain high resolution, it also provides a unique opportunity to study the conformational flexibility/dynamics of the macromolecular assemblies. Ideally, homogenous samples have to be biochemically standardized and prepared before the vitrification process. However, this is not possible with all protein samples due to the inherent protein flexibility which is necessary for its function, for example, rotation of 30S subunit of ribosome [106] or rotational states in case of eukaryotic V-ATPase [107]. In such cases, the heterogeneous sample data images can be classified computationally to classes containing homogenous particles (an example of such classification can be seen in Fig. 4).
Three main techniques are currently in use to identify and sort the macromolecular structural conformational variability or heterogeneity [41]. The first approach depends on classifying the 2D images based on the eigen images/eigenvectors [74, 108,109,110,111] without any starting model. First, classify using MSA to obtain orientation classes, and then, the major variation among the picked particles in each orientation classes can be identified in the low-order eigen images by MSA, and using these, information particles can be classified into homogenous classes, leading to preliminary 3D reconstruction from a class containing majority of homogenous particles as shown in Fig. 3. The preliminary 3D reconstructions can be projected as references for competitive alignment. Further, the quality of 3D reconstruction can be iteratively improved until the eigen images show no major variations within the class and the particles stabilize from jumping to another class during competitive projection matching. In this manner, three class reconstructions were obtained as shown in Fig. 4. The second method depends on detection in 2D variations using starting model [41]. The third method also needs initial starting model and uses a statistical approach to obtain 3D classification. In this case, large number of 3D maps are calculated from randomly selected subset of particles (with previously assigned orientation based on initial 3D map). Determination of the 3D variance can be used to assess the heterogeneity , and estimation of covariance enables one to carry out the 3D classification according to variable regions. Alternatively, the molecular states can be separated using maximum likelihood classification [104, 112] or by the latest multi-body refinement method [113].
5 Single-Particle Cryo-EM Applications in SBDD
There are five 3D reconstructions in the EMDB with bound inhibitors or ligands at resolution better than 2.5 Å as shown in Table 1. We have focused at this resolution since this is at the center of medium (3.0 Å) and high resolution (1.8 Å), which is desired resolution for SBDD studies. Although we have highlighted reconstructions better than 2.5 Å, ligands have been visualized in the 3D reconstructions better than 4.0 Å. While there are only 10s of EM reconstructions with bound inhibitors at 2.5 Å or better, there are several 100s of structures in the EMDB at resolution between 2.5 and 4 Å with bound inhibitors or ligands. Here are couple of examples of 3D reconstructions with resolution better than 2.5 Å: In the Sect. 2.3 we have already come across the example of 3D reconstruction (by the Subramaniam group [75]) of the inhibitor PETG bound to beta-galactosidase at 1.9 Å resolution. His group used a similar approach to solve the cryo-EM structure of human p97ATPase, an important target for cancer, in complex with its allosteric inhibitor UPCDC30245 [10] as shown in Fig. 7c . Although they could not see a part of the inhibitor in the EM electron potential map, they could see at 2.3 Å resolution the other part where the inhibitor snugly fits into the protein pocket and proposed how the allosteric inhibitor UPCDC30245 inhibits the conformational changes necessary for the function of p97. They further could see three coexisting functional states of p97 in the presence and absence of ATPγS. Here are couple of examples of 3D reconstructions of proteins bound to inhibitors, with 3D reconstruction resolution poorer than 2.5 Å. For example, Paula and Ed solved the structure of 20S proteasome in complex with the inhibitor (EMD-3231) at ~3.6 Å resolution [114] as shown in Fig. 7a . Another example is the structure of 70S ribosome from Escherichia coli at 2.9 Å resolution in complex with elongation factor Tu, aminoacyl-tRNA, and the antibiotic kirromycin [115] as shown in Fig. 7b . With these examples, it is very clear that, in the future, the single-particle cryo-EM will play a very important role in the preclinical SBDD studies.
a Inhibitor phenylethyl β-D-thiogalactopyranoside (PETG) (blue surface)-bound cryo-EM structure of β-galactosidase enzyme at 1.9 Å resolution [75]. b Zoom-in view of the squared area with bound inhibitor PETG (blue surface). The EM map is shown in yellow mesh. Sodium (magenta) and Mg2+ (green) ions and water molecules (red) can be seen in the pocket
Pharmacologically important target proteins (beige color) (modeled using the cryo-EM electron potential 3D reconstruction map) in complex with inhibitor (shown as stick model). The cryo-EM map of the inhibitors are shown as blue mesh. a 3.6 Å reconstruction of 20S Plasmodium falciparum proteasome [114] with bound inhibitor Mor-WLW vinyl sulfone (EMD3231) (PDB ID: 5fmg). b 2.9 Å cryo-EM reconstruction of complete 70S Escherichia coli ribosome with bound antibiotic kirromycin [115] (EMD 2847, PDB ID: 5afi). c 2.3 Å resolution 3D reconstruction of anticancer drug target human p97 with bound allosteric inhibitor UPCDC30245 [10]
6 Conclusions and Future Prospective
Recent advances in cryo-EM have enabled us to use single-particle cryo-EM as a method of choice to resolve solution-state 3D structures of proteins and protein complexes at atomic resolution, thus breaking the cryo-EM resolution barrier to facilitate SBDD [6]. In recent years, many pharmaceutical companies like Bayer, Merck Research Laboratories, Sonafi, AstraZeneca, Regeneron Pharmaceuticals, NovAliX, Genentech etc. have realized the importance of this method and started hiring experts in single-particle cryo-EM to get involved in their SBDD pipeline. Table 1 lists the protein structures with bound ligands solved by single particle cryo-EM at resolution 2.5 Å or better; i.e., five of the structures have bound inhibitors/glycans, which underscore the importance of single-particle cryo-EM in SBDD. Apart from these, there are many more structures with bound ligands in the EMDB at resolutions below 2.5 Å. The future of this technique will be in obtaining the sub-nanometer resolution and perhaps atomic-resolution structures of proteins and protein complexes in vivo. This methodology called the cellular tomography, although not the scope of this chapter, is a promising future technology for atomic-resolution structures of proteins and protein complexes in its native environment “the cell.” With the advent of phase plates, energy filters, and automation in cryo-EM data collection, promising efforts are being made to achieve that goal and the realization of that goal may not be far away which would in turn potentially further accelerate the SBDD program.
Abbreviations
- 3D:
-
Three Dimension
- CTF:
-
Contrast Transfer Function
- cryo-EM:
-
Cryo-electron microscopy
- CC:
-
Cross-Correlation
- DDD or DED:
-
Direct Detection Device or Direct Electron Detector
- ET:
-
Electron Tomography
- EMDB:
-
Electron Microscopy Data Bank
- EM:
-
Electron Microscopy
- FEG:
-
Field Emission Gun
- FSC:
-
Fourier Shell Correlation
- MSA:
-
Multivariate Statistical Analysis
- PDB:
-
Protein Data Bank
- PCA:
-
Principle Component Analysis
- SBDD:
-
Structure-Based Drug Design
- SNR:
-
Signal-to-Noise Ratio
- SSNR:
-
Spectral SNR
- TEM:
-
Transmission Electron Microscopy
References
Dauter Z, Lamzin VS, Wilson KS (1995) Proteins at atomic resolution. Curr Opin Struct Biol 5:784–790
Kozma D, Simon I, Tusnady GE (2013) PDBTM: protein data bank of transmembrane proteins after 8 years. Nucleic Acids Res 41:D524–D529
Natesh R (2014) Crystallography beyond crystals: PX and SP cryoEM. Resonance 19:1177–1196
Mountain V (2003) Astex, structural genomix, and syrrx. i can see clearly now: structural biology and drug discovery. Chem Biol 10:95–98
Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S (2015) A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 348:1147–1151
Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, Subramaniam S (2016) Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165:1698–1707
Shalev-Benami M, Zhang Y, Rozenberg H, Nobe Y, Taoka M, Matzov D, Zimmerman E, Bashan A, Isobe T, Jaffe CL, Yonath A, Skiniotis G (2017) Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat Commun 8:1589
Dong Y, Liu Y, Jiang W, Smith TJ, Xu Z, Rossmann MG (2017) Antibody-induced uncoating of human rhinovirus B14. In: Proceedings of the national academy of sciences of the United States of America, vol 114, pp 8017–8022
Danev R, Tegunov D, Baumeister W (2017) Using the Volta phase plate with defocus for cryo-EM single particle analysis. Elife 6:1–9
Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JL, Huryn D, Arkin M, Subramaniam S (2016) A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351:871–875
Dubochet J, Mcdowall AW (1981) Vitrification of pure water for electron-microscopy. J Microsc Oxford 124:Rp3–Rp4
Vogel RH, Provencher SW, von Bonsdorff CH, Adrian M, Dubochet J (1986) Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature 320:533–535
Henderson R, Unwin PN (1975) Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257:28–32
Frank J (1975) Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1:159–162
Frank J, Al-Ali L (1975) Signal-to-noise ratio of electron micrographs obtained by cross correlation. Nature 256:376–379
Saxton WO, Frank J (1977) Motif detection in quantum noise-limited electron micrographs by cross-correlation. Ultramicroscopy 2:219–227
Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH (1990) Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol 213:899–929
Khoshouei M, Radjainia M, Baumeister W, Danev R (2017) Cryo-EM structure of haemoglobin at 3.2 A determined with the Volta phase plate. Nat Commun 8:16099
Henderson R (1995) The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys 28:171–193
Yu X, Jin L, Zhou ZH (2008) 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453:415–419
Zhang X, Sun S, Xiang Y, Wong J, Klose T, Raoult D, Rossmann MG (2012) Structure of Sputnik, a virophage, at 3.5 Å resolution. Proc Nat Acad Sci USA 109:18431–18436
Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W (2011) Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Nat Acad Sci USA 108:1355–1360
Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P (1988) Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21:129–228
Passmore LA, Russo CJ (2016) Specimen preparation for high-resolution cryo-EM. Methods Enzymol 579:51–86
Russo CJ, Passmore LA (2016) Progress towards an optimal specimen support for electron cryomicroscopy. Curr Opin Struct Biol 37:81–89
Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y (2013) Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590
McMullan G, Clark AT, Turchetta R, Faruqi AR (2009) Enhanced imaging in low dose electron microscopy using electron counting. Ultramicroscopy 109:1411–1416
Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332
Abrishami V, Vargas J, Li X, Cheng Y, Marabini R, Sorzano CO, Carazo JM (2015) Alignment of direct detection device micrographs using a robust optical flow approach. J Struct Biol 189:163–176
de la Rosa-Trevin JM, Oton J, Marabini R, Zaldivar A, Vargas J, Carazo JM, Sorzano CO (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J Struct Biol 184:321–328
Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, Potter CS, Carragher B, Henderson R, Grigorieff N (2012) Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol 177:630–637
Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, Grigorieff N (2012) Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20:1823–1828
Russo CJ, Passmore LA (2014) Electron microscopy: ultrastable gold substrates for electron cryomicroscopy. Science 346:1377–1380
Russo CJ, Passmore LA (2014) Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nat Methods 11:649–652
Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60
Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51
Li X, Zheng S, Agard DA, Cheng Y (2015) Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol 192:174–178
Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh MP, Booth CR, Shinkawa T, Nakata M, Chiu W (2009) JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J Struct Biol 165:1–9
Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B (2009) Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102
Hoppe W (1983) Electron-diffraction with the transmission electron-microscope as a phase-determining diffractometer—from spatial-frequency filtering to the 3-Dimensional structure-analysis of ribosomes. Angew Chem Int Edit 22:456–485
Orlova EV, Saibil HR (2011) Structural analysis of macromolecular assemblies by electron microscopy. Chem Rev 111:7710–7748
Erickson HP, Klug A (1971) Measurement and compensation of defocusing and aberrations by fourier processing of electron micrographs. Philos T Roy Soc B. 261:105–118
Wade RH (1992) A brief look at imaging and contrast transfer. Ultramicroscopy 46:145–156
Bell JM, Chen M, Baldwin PR, Ludtke SJ (2016) High resolution single particle refinement in EMAN2.1. Methods 100:25–34
Rohou A, Grigorieff N (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221
Zhang K (2016) Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12
Mallick SP, Carragher B, Potter CS, Kriegman DJ (2005) ACE: automated CTF estimation. Ultramicroscopy 104:8–29
Roseman AM (2004) FindEM–a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol 145:91–99
Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46
Ludtke SJ, Bell JM, Chen M, Baldwin PR, Ludtke SJ (2016) Single-particle refinement and variability analysis in EMAN2.1, high resolution single particle refinement in EMAN2.1. Methods Enzymol 579:159–189
van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116:17–24
Smith JM (1999) Ximdisp–a visualization tool to aid structure determination from electron microscope images. J Struct Biol 125:223–228
Wood C, Burnley T, Patwardhan A, Scheres S, Topf M, Roseman A, Winn M (2015) Collaborative computational project for electron cryo-microscopy. Acta Crystallogr D Biol Crystallogr 71:123–126
Burnley T, Palmer CM, Winn M (2017) Recent developments in the CCP-EM software suite. Acta Crystallogr Sect D Struct Biol 73:469–477
Scheres SH (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530
Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296
Heimowitz A, Andén J, Singer A (2018) APPLE picker: automatic particle picking, a low-effort Cryo-EM framework. J Struct Biol 204(2):215–227
Singer A et al. (2010) Algorithms for Single Particle Reconstruction (ASPIRE), http://spr.math.princeton.edu/
Hoang TV, Cavin X, Schultz P, Ritchie DW (2013) gEMpicker: a highly parallel GPU-accelerated particle picking tool for cryo-electron microscopy. BMC Struct Biol 13:25
Chen JZ, Grigorieff N (2007) SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol 157:168–173
Henderson R (2013) Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proc Nat Acad Sci USA 110:18037–18041
De Rosier DJ, Klug A (1968) Reconstruction of three dimensional structures from electron micrographs. Nature 217:130–134
Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116:190–199
Zivanov J, Nakane T, Scheres S (2018) A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis, bioRxiv
Lyumkis D, Brilot AF, Theobald DL, Grigorieff N (2013) Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol 183:377–388
de la Rosa-Trevin JM, Quintana A, Del Cano L, Zaldivar A, Foche I, Gutierrez J, Gomez-Blanco J, Burguet-Castell J, Cuenca-Alba J, Abrishami V, Vargas J, Oton J, Sharov G, Vilas JL, Navas J, Conesa P, Kazemi M, Marabini R, Sorzano CO, Carazo JM (2016) Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J Struct Biol 195:93–99
Frank J, Shimkin B, Dowse H (1981) Spider—a modular software system for electron image processing. Ultramicroscopy 6:343–357
van Heel M, Frank J (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6:187–194
Scheres SH (2012) A Bayesian view on cryo-EM structure determination. J Mol Biol 415:406–418
Van Heel M (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21:111–123
Penczek PA, Grassucci RA, Frank J (1994) The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 53:251–270
Radermacher M, Wagenknecht T, Verschoor A, Frank J (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146:113–136
Penczek PA (2014).http://sparx-em.org/sparxwiki/sxviper
Natesh R, Clare DK, Farr GW, Horwich AL, Saibil HR (2018) A two-domain folding intermediate of RuBisCO in complex with the GroEL chaperonin. Int J Biol Macromol 118:671–675
Bartesaghi A, Aguerrebere C, Falconieri V, Banerjee S, Earl LA, Zhu X, Grigorieff N, Milne JLS, Sapiro G, Wu X, Subramaniam S (2018) Atomic resolution cryo-EM structure of beta-galactosidase. Structure 26(848–856):e3
Penczek PA (2010) Resolution measures in molecular electron microscopy. Methods Enzymol 482:73–100
Harauz G, van Heel M (1986) Exact filters for general geometry three-dimensional reconstruction. Optik 73:146–156
van Heel M, Schatz M (2005) Fourier shell correlation threshold criteria. J Struct Biol 151:250–262
Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333:721–745
Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroder GF, Steven AC, Stokes DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL (2012) Outcome of the first electron microscopy validation task force meeting. Structure 20:205–214
Kucukelbir A, Sigworth FJ, Tagare HD (2014) Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65
Cardone G, Heymann JB, Steven AC (2013) One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol 184:226–236
Natesh R, Manikandan K, Bhanumoorthy P, Viswamitra MA, Ramakumar S (2003) Thermostable xylanase from Thermoascus aurantiacus at ultrahigh resolution (0.89 A) at 100 K and atomic resolution (1.11 A) at 293 K refined anisotropically to small-molecule accuracy. Acta Crystallogr D Biol Crystallogr 59:105–117
Natesh R, Bhanumoorthy P, Vithayathil PJ, Sekar K, Ramakumar S, Viswamitra MA (1999) Crystal structure at 1.8 A resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus. J Mol Biol 288:999–1012
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Jones TA (2004) Interactive electron-density map interpretation: from INTER to O. Acta Crystallogr D Biol Crystallogr 60:2115–2125
Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G (2016) Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr 71:136–153
Echols N, Moriarty NW, Klei HE, Afonine PV, Bunkoczi G, Headd JJ, McCoy AJ, Oeffner RD, Read RJ, Terwilliger TC, Adams PD (2014) Automating crystallographic structure solution and refinement of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 70:144–154
Baker ML, Baker MR, Hryc CF, Ju T, Chiu W (2012) Gorgon and pathwalking: macromolecular modeling tools for subnanometer resolution density maps. Biopolymers 97:655–668
Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A (2008) Protein structure fitting and refinement guided by cryo-EM density. Structure 16:295–307
Joseph AP, Malhotra S, Burnley T, Wood C, Clare DK, Winn M, Topf M (2016) Refinement of atomic models in high resolution EM reconstructions using Flex-EM and local assessment. Methods 100:42–49
Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K (2009) Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49:174–180
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(33–8):27–28
Schrodinger LLC (2015) The PyMOL molecular graphics system, Version 1.8 in
Brunger AT (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–475
Shaikh TR, Hegerl R, Frank J (2003) An approach to examining model dependence in EM reconstructions using cross-validation. J Struct Biol 142:301–310
Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R (2013) High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135:24–35
Rosenthal PB, Rubinstein JL (2015) Validating maps from single particle electron cryomicroscopy. Curr Opin Struct Biol 34:135–144
Scheres SH, Chen S (2012) Prevention of overfitting in cryo-EM structure determination. Nat Methods 9:853–854
Murray SC, Flanagan J, Popova OB, Chiu W, Ludtke SJ, Serysheva II (2013) Validation of cryo-EM structure of IP(3)R1 channel. Structure 21:900–909
Farabella I, Vasishtan D, Joseph AP, Pandurangan AP, Sahota H, Topf M (2015) TEMPy: a Python library for assessment of three-dimensional electron microscopy density fits. J Appl Crystallogr 48:1314–1323
Henderson R, Chen S, Chen JZ, Grigorieff N, Passmore LA, Ciccarelli L, Rubinstein JL, Crowther RA, Stewart PL, Rosenthal PB (2011) Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. J Mol Biol 413:1028–1046
Scheres SH (2010) Classification of structural heterogeneity by maximum-likelihood methods. Methods Enzymol 482:295–320
Elmlund D, Le SN, Elmlund H (2017) High-resolution cryo-EM: the nuts and bolts. Curr Opin Struct Biol 46:1–6
Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003) Locking and unlocking of ribosomal motions. Cell 114:123–134
Zhao J, Benlekbir S, Rubinstein JL (2015) Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 521:241–245
White HE, Orlova EV, Chen S, Wang L, Ignatiou A, Gowen B, Stromer T, Franzmann TM, Haslbeck M, Buchner J, Saibil HR (2006) Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. Structure. 14:1197–1204
White HE, Saibil HR, Ignatiou A, Orlova EV (2004) Recognition and separation of single particles with size variation by statistical analysis of their images. J Mol Biol 336:453–460
Elad N, Clare DK, Saibil HR, Orlova EV (2008) Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J Struct Biol 162:108–120
Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR (2007) Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell 26:415–426
Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM (2007) Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4:27–29
Nakane T, Kimanius D, Lindahl E, Scheres SH (2018) Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife. 7:1–18
da Fonseca PC, Morris EP (2015) Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat Commun 6:7573
Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H (2015) Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520:567–570
Acknowledgements
RN was supported by Ramalingaswamy Fellowship from DBT. RN would like to thank his laboratory members and colleagues for their constant support, valuable scientific and technical discussion. Last but not least, RN would like to thank IISER-TVM past Director Prof. ED Jemmis and present Director Prof. V. Ramakrishnan for their unstinted support.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Natesh, R. (2019). Single-Particle cryo-EM as a Pipeline for Obtaining Atomic Resolution Structures of Druggable Targets in Preclinical Structure-Based Drug Design. In: Mohan, C. (eds) Structural Bioinformatics: Applications in Preclinical Drug Discovery Process. Challenges and Advances in Computational Chemistry and Physics, vol 27. Springer, Cham. https://doi.org/10.1007/978-3-030-05282-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-05282-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-05281-2
Online ISBN: 978-3-030-05282-9
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)