Abstract
In this project, our goal is to classify different types of liver tissue on 3D multi-parameter magnetic resonance images in patients with hepatocellular carcinoma. In these cases, 3D fully annotated segmentation masks from experts are expensive to acquire, thus the dataset available for training a predictive model is usually small. To achieve the goal, we designed a novel deep convolutional neural network that incorporates auto-context elements directly into a U-net-like architecture. We used a patch-based strategy with a weighted sampling procedure in order to train on a sufficient number of samples. Furthermore, we designed a multi-resolution and multi-phase training framework to reduce the learning space and to increase the regularization of the model. Our method was tested on images from 20 patients and yielded promising results, outperforming standard neural network approaches as well as a benchmark method for liver tissue classification.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Tissue classification
- Convolutional neural network
- Auto-context
- Multi-phase training
- Hepatocellular carcinoma
- Magnetic resonance imaging
1 Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancer types and the leading cause in cancer-related death [4]. Multi-parameter dynamic contrast enhanced (DCE) magnetic resonance (MR) images are commonly used as a diagnostic tool for suspected HCC cases and are important for defining treatment targets and predicting outcomes for a number of therapeutic strategies including transarterial chemoembolization (TACE) [3]. In this work, we are interested in classifying liver tissue into clinically relevant types on 3D MR images: parenchyma and anomalies that consist of viable tumor tissue and necrosis tissue. Recent developments in the design of deep convolutional neural networks (CNN) provide ways to construct powerful models that can extract both low and high level features from images that are usually difficult to formulate with traditional methods and draw accurate inferences [5]. However, such models typically need a large amount of expert curated labels. This is particularly expensive in our case as the training requires 3D fully annotated segmentation masks from radiologists.
To overcome these challenges, we designed a novel CNN model that incorporates contextual information to perform classification in a local patch region. The input patches were sampled at a fixed size but with different resolutions, in order to capture information from different scales efficiently. We developed an auto-context-based multi-level architecture that, when coupled with a multi-phase training procedure, can effectively learn and predict at different levels. The learning space needed for the each level of the model was thus reduced, since it only needed to learn the incremental difference based on the learner in the previous level.
Several other works have explored the similar idea of combining CNN and auto-context [6, 9]. Here we want to point out the difference. In a popular study [6], auto-context is applied outside the classifier to refine classification performance. Our algorithm, in contrast, applies auto-context within the multi-level classifier, efficiently integrating contextual information from multi-resolution patch samples to address the small dataset problem.
The main contributions of this work are threefold: (1) It is the first deep neural network approach to segment tissue types on multi-parameter MR images in HCC patients without the need of manually designing image features [7]. While deep CNNs have been developed for liver tumor segmentation from CT images [1, 2], such approaches have not been applied to MR images. (2) It incorporates a novel auto-context based CNN model design combined with a multi-phase training strategy that encourages the model to utilize contextual information from the previous phase. This hierarchical combination of several predictive units is shown to out-perform the use of a single U-net model given the available data pool without overfitting. (3) It creatively addresses the data deficiency problem by sampling the image at different resolutions under a patch based learning scheme. These multi-resolution patches effectively integrate image information from different scales yet maintain a relatively low input dimensionality. Overall, we see the methodology employed in this work as being generalizable to a number of other detection and segmentation tasks in biomedical images where full image annotation is difficult to acquire.
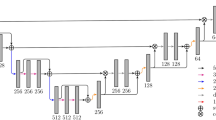
Overall structure. Subfigure (a) illustrates the overall architecture of the model. \(x^{(k)}\)’s are the patches sampled from the image at resolution k’s, \(y^{(k)}\)’s are the corresponding output of each unit k. \(m^{(k)}\)’s are different sizes of Gaussian-shape masks applied to \(y^{(k)}\)’s to emphasize prediction performance at the center of patches. Dashed lines between \(x^{(k)}\)’s and the units means connections are optional. Subfigure (b) illustrates the sampling patterns at different resolutions: the same window dimension, but different voxel-to-voxel distance
2 Proposed Method
2.1 Data Preprocessing
We adopted a patch-based learning scheme in our study to address the data deficiency problem, as the model would only need to learn the probability distribution of each voxel at a local patch region. In addition, we designed a weighted sampling procedure to address the class imbalance problem. On average, anomalies account for only 10% of the total liver tissue. We thus re-balanced the class by forcing a sampling frequency of 50% parenchyma and 50% anomalies.
We also implemented a novel multi-resolution sampling procedure to incorporate image information at different scales in each patch. This is useful for detecting and delineating anomalies at different sizes (Fig. 1a). This multi-resolution sampling method has two advantages over simply expanding the patch size with a fixed resolution. First, the fixed patch size is more convenient to work with in CNNs. Second, the number of voxels in the input array is greatly reduced to improve computation efficiency.
To further handle the small dataset problem, we used data augmentation. Each time a patch was sampled, a 3D random rotation was applied.
2.2 Multi-level Hierarchical Architecture
The architecture we proposed is illustrated in Fig. 1b. The whole model consists of three basic units. In general, each unit k can be any CNN that outputs a probability map, but in this study we adopted the U-net architecture due to its elegant design and powerful performance [5]. The entire model took in image patches sampled at different resolutions and output predictions at those resolutions. The connection from output \(y^{k}\) from each unit to its higher level unit draws inspiration from the research in auto-context [8].
We used a weighted cross entropy as our loss function to update the weights in the neural network (Eq. 1), and a weighted dice similarity coefficient to monitor the training process and to select the best model (Eq. 2).
In Eqs. (1) and (2), x is the location inside the patch, i is the class, p is the true probability distribution, taking only values of 0 or 1, q is the predicted probability distribution, m is a Gaussian shape mask to emphasize the performance at the center of the patch, \(\omega \) and \(\alpha \) are the weights in the loss function and the metric that are set to accentuate performance on certain classes, and \(\varOmega _{h, i}\) is the segmentation mask for class i based on a probability map h.
2.3 Multi-phase Training Procedure
During the training process, the model was trained in three coarse-to-fine phases. For example, in the first phase of training, weights in unit 3 were updated, while weights in unit 2 and 1 were frozen; then in the second phase of training, weights in unit 2 were updated, while those in unit 3 and 1 were frozen. This multi-phase training procedure was employed to reduce the risk of overfitting for the whole model and it was based on our intuition that the output of each unit should function as a coarse estimation at its resolution. This regularization is helpful in our case for two reasons: (1) Our image data pool is limited even with random sampling and rotation-based data augmentation. (2) The ground truth is not necessarily reliable as manual segmentation in noisy 3D images is prone to errors. Similar methodology has been reported in several recent works [10].
2.4 Data Postprocessing
During the prediction step, the predicted probability map for the whole image was assembled together by summing all predicted patches with overlap while each patch is weighted by a Gaussian mask as specified in Eq. 1, since the model was trained to emphasize the performance at the center of the patch. Simple post processing was used to get rid of small anomalies in the predicted masks by setting the label of those anomalies whose volume were under a certain threshold to parenchyma.
Segmentation demonstration. Red color is the parenchyma, green color is the viable tumor tissue, blue color is the necrosis. Subfigure 2a shows an expert delineation of some viable tumor tissue and necrosis. Subfigures 2b to d show the prediction results from three other methods, namely single-resolution input single-phase training, multi-resolution input single-phase training, and the benchmark method, manually designed features with random forest in auto-context, as described in Sect. 3.1. Subfigures 2e to g show the three-phase coarse-to-fine prediction progression in the proposed method. Visualization is provided using the software itk-SNAP. (Color figure online)
3 Experiments and Results
3.1 Experiment Setup
The image data we used included 20 sets of multi-parameter 3D MR images, each of which consisted of one T2 weighted MR image and three T1 weighted dynamic enhanced contrast images at three different time points during the surgical intervention: pre-contrast phase (before the contrast injection), arterial phase (20 s after the injection), and venous phase (70 s after the injection). All four images were mutually registered. Though a full automation that included liver segmentation was possible under our framework, liver masks were provided in order to achieve a fair comparison with the benchmark method, and to focus on the problem of the delineation inside the liver. Each patient’s image intensity was normalized to roughly between 0 and 1.
Images used in this study are from HCC patients with TACE procedures as part of a larger clinical study on treatment outcome analysis. In these cases, the number of anomalies often ranges from 1 to 3, with diameter over 20 mm. During the TACE procedure, the largest tumors are the most important targets. Therefore the resolutions were selected as 2 mm, 1 mm and 1 mm, with a patch size of 16-by-16-by-16 voxels, in order to focus on performance on medium and large size tumors. The 20-patient dataset generated effectively 1700 non-overlapping patches, though with random sampling and random rotation augmentation, no patches would be exactly the same.
The first two units of the model were designed to differentiate anomalies from normal liver tissue, while the last one was designed to identify viable tumor tissue inside each detected anomaly. This was done by tuning the class weight \(\omega \) in the loss function (Eq. 1). In phase 1, the \(\omega \)’s for parenchyma, viable tumor tissue, and necrosis are (1.0, 2.0, 0.3), phase 2 (1.0, 1.5, 0.3), and phase 3 (0.0, 1.0, 2.0). For each unit in the model, we implemented a U-net CNN with ten layers of \(3\times 3\times 3\) convolution, ten layers of dropout, and two levels of max-pooling/upsampling. Five fold cross validation method was used to evaluate the performance of different models. Hyperparameters, such as learning rate and class weights in the loss functions, remained the same across all five folds.
3.2 A Combination of Measurements
In our evaluation of the method, we also included a two-step measurement instead of solely the traditional dice similarity coefficient (DSC). First, we calculated how well the anomalies were detected using F score (Eq. 3).
We set \(\beta =2\) to reflect the emphasis on recall rate in a clinical setting. An anomaly is detected if part of its voxels are covered by some predicted masks. Second, we measured how good the delineation was by aggregating all regions of interest (anomalies and viable tumor tissue) together and calculating the DSC. We provide a toy example to further explain the difference between the detection metric and the delineation metric in Fig. 3.
Examples of difference between detection and delineation. Blue regions stand for anomalies. Orange regions stand for predictions. Subfigure (a): good delineation (high DSC), poor detection (low F score). Subfigure (b): medium delineation, good detection. Subfigure (c): poor delineation, good detection. (Color figure online)
3.3 Results
Figure 2 demonstrates an example of the proposed algorithm output. Table 1 summarizes the results in our study. The different rows in the method column describe whether the model utilized multi-resolution input, or only the resolution at the lowest level; whether it trained the model with a multi-phase strategy, or without. The single-resolution input single-phase training method is equivalent to the traditional U-net method. The benchmark method uses manually designed image features with random forest and iteratively trained auto-context classifiers as described in [7]. Figure 4 describes how well the different models delineate anomalies at different sizes.
We make several observations from the results we present here.
-
1.
The proposed method achieved the best overall anomaly and viable tumor tissue delineation performance, compared to both other CNN-based methods and the benchmark method.
-
2.
The proposed method was tuned towards and did achieve the best performance in delineating medium and large size anomalies which the TACE procedure was targeting.
-
3.
The proposed method was highly efficient in implementation. The whole model was trained within 90 min without the need of manually designing complex image features, while it took 18 hours for the benchmark method to finish running on a better machine.
4 Conclusion
In this work we presented a deep neural network approach to detect and delineate different types of liver tissue on multi-parameter MR images in patients with HCC. The patch-based algorithm was able to achieve a performance level that was better than the benchmark method without the need of manually designing different shape and texture features, with an implementation that was much more efficient. The multi-resolution input, the auto-context design and the multi-phase training procedure were helpful in improving overall performance compared to the traditional U-net architecture. In the future, this method can be applied to a full delineation of the liver tissue with any number of hierarchical tissue types, including the liver itself. In addition, this methodology can be applied to a number of other detection and delineation problems in the biomedical imaging field.
References
Christ, P.F., et al.: Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 415–423. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_48
Li, W., Jia, F., Hu, Q.: Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J. Comput. Commun. 3(11), 146 (2015)
Raoul, J.L., et al.: Evolving strategies for the management of intermediate-stage hepatocellular carcinoma: available evidence and expert opinion on the use of transarterial chemoembolization. Cancer Treat. Rev. 37(3), 212–220 (2011)
Raza, A., Sood, G.K.: Hepatocellular carcinoma review: current treatment, and evidence-based medicine. World J. Gastroenterol. WJG 20(15), 4115 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Salehi, S.S.M., Erdogmus, D., Gholipour, A.: Auto-context convolutional neural network (auto-net) for brain extraction in magnetic resonance imaging. IEEE Trans. Med. Imaging 36(11), 2319–2330 (2017)
Treilhard, J., et al.: Liver tissue classification in patients with hepatocellular carcinoma by fusing structured and rotationally invariant context representation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 81–88. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_10
Tu, Z., Bai, X.: Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 32(10), 1744–1757 (2010)
Vodopivec, T., Lepetit, V., Peer, P.: Fine hand segmentation using convolutional neural networks. CoRR abs/1608.07454 (2016). http://arxiv.org/abs/1608.07454
Zeng, G., Yang, X., Li, J., Yu, L., Heng, P.-A., Zheng, G.: 3D U-net with multi-level deep supervision: fully automatic segmentation of proximal femur in 3D MR images. In: Wang, Q., Shi, Y., Suk, H.-I., Suzuki, K. (eds.) MLMI 2017. LNCS, vol. 10541, pp. 274–282. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67389-9_32
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, F. et al. (2018). Liver Tissue Classification Using an Auto-context-based Deep Neural Network with a Multi-phase Training Framework. In: Bai, W., Sanroma, G., Wu, G., Munsell, B., Zhan, Y., Coupé, P. (eds) Patch-Based Techniques in Medical Imaging. Patch-MI 2018. Lecture Notes in Computer Science(), vol 11075. Springer, Cham. https://doi.org/10.1007/978-3-030-00500-9_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-00500-9_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00499-6
Online ISBN: 978-3-030-00500-9
eBook Packages: Computer ScienceComputer Science (R0)