Abstract
Although atypical bacteria are important causes of lower airway infections, data on their role in immunocompromised patients are scarce. The aim of the study was to evaluate the prevalence of atypical pulmonary infections in patients with various types of immunosuppression, and to analyze clinical characteristics of these infections. Eighty non-HIV immunocompromised patients with different underlying diseases and clinical and radiological signs of pulmonary infection were enrolled. Due to incomplete data on eight patients, 72 patients were eligible for final analysis (median age 58 years). All patients underwent fiberoptic bronchoscopy and bronchoalveolar lavage. Bronchoalveolar lavage fluid (BALF) fluid samples were sent for direct microscopy, cultures, and fungal antigen detection, when appropriate. Commercial qualitative amplification assay (PNEUMOTRIS oligomix Alert Kit®), based on nested PCR method, was used to detect specific DNA sequences of L. pneumophila, C. pneumoniae, and M. pneumoniae in BALF. There were 61 (84.7 %) patients with hematologic diseases, 3 (4.2 %) after solid organ transplantation, and 8 (11.1 %) with miscellaneous diseases affecting immune status. Specific sequences of M. pneumoniae, C. pneumoniae, and L. pneumophila DNA were found in 7 (9.7 %), 2 (2.8 %), and 0 patients, respectively. In 8 of these patients co-infections with different microorganisms were demonstrated. Co-infection with A. baumanii and P. aeruginosa was diagnosed in three patients who died. We conclude that atypical lower airway infections are uncommon in immunocompromised patients. The majority of these infections are co-infections rather than single pathogen infections.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Atypical bacteria
- Bronchoalveolar lavage fluid
- Chlamydophila pneumoniae
- Legionella pneumophila
- Mycoplasma pneumoniae
- Immunodeficiency
- Respiratory infections
1 Introduction
The incidence of lower airway infections in immunocompromised patients is high and the course of a disease is usually more severe than that in immunocompetent hosts (Sousa et al. 2013; Bonatti et al. 2009). Mortality rate largely depends on the type and severity of immunosuppression, with the highest rate reported after hematopoietic stem cell transplantation (HSCT) and somewhat lower in solid organ transplant (SOT) recipients and patients with hematologic malignancies (HM) (Cervera et al. 2006; Rañó et al. 2001; 2002). It has also been shown that the outcome of pulmonary infections is significantly affected by a delay in diagnosis of specific etiology. An increase in mortality rate from 29 to 71 % has been reported in patients in whom the etiology of infection was ascertained within the first 7 days of onset of symptoms compared with those with later diagnosis (Rañó et al. 2001). The etiology of lower respiratory tract infections in immunocompromised patients is diverse. It includes common bacteria, uncommon bacterial agents, and opportunistic pathogens such as various fungal species and viruses. Although atypical bacteria are important causes of pulmonary infections in the general population, data on the role of these pathogens in immunocompromised patients are relatively scarce. In the immunocompetent hosts Mycoplasma pneumoniae and Chlamydophila pneumoniae are responsible for 1–36 % and 3–22 % of community acquired pneumonia (CAP) cases, respectively (Singanayagam et al. 2014; Masiá et al. 2007; Gleason 2002). The majority of these infections affect children and young adults and present as mild, self-limiting disease (Capelastegui et al. 2012). However, even 26 % of patients may require hospital admission and in-hospital death rate may be as high as 5 %. The prevalence of Legionella pneumophila pneumonia in the general population is slightly lower (2–16 %), but this infection is usually more severe. In two studies, L. pneumophila was responsible for 2–9 % of CAP that required hospitalization (Yu and Stout 2008; Gleason 2002). On the other hand, recent data do not confirm the relation between L. pneumophila infection and increased in-hospital mortality rate (Capelastegui et al. 2012).
It might be hypothesized that the course of atypical pulmonary infections in immunocompromised patients can be more severe than that in the general population and that the co-infection with atypical pathogens can aggravate the course of pulmonary disease caused by typical bacteria or fungi. Surprisingly, there are little data on the incidence and clinical features of atypical pulmonary infections in immunocompromised patients. According to the available publications, the incidence of these infections is quite low (Corti et al. 2009; Jain et al. 2004; Perez and Leigh 1991). However, a few cases of life threatening pneumonia caused by C. pneumoniae and L. pneumophila have been described (Di Stefano et al. 2007; Heinemann et al. 2000). Whether the true prevalence of atypical pathogen infections in immunocompromised hosts is low or it is underestimated due to low sensitivity of the diagnostic methods seems to be an interesting issue. It must be realized that the culture of atypical bacteria is difficult and demanding and can be offered by few laboratories only. Serological methods, including specific IgM and IgG antibodies detection in the serum, have limited clinical application due to a delay in the diagnosis and suboptimal sensitivity in patients with immunoglobulin deficiency (false negative results) (Bartlett 2008; Hammerschlag 2000; Welti et al. 2003). Likewise, L. pneumophila antigen detection in the urine has limited sensitivity as a negative result of this test does not exclude infection with other than serotype 1 L. pneumophila strains. The introduction of polymerase chain reaction (PCR)-based methods that can identify specific genetic material in different biological samples, including broncholaveolar lavage fluid (BALF), throat swabs, and nasopharyngeal samples, enables a rapid, sensitive, and specific diagnosis of atypical pathogen infection even if patients are already treated with an antibiotic (Murdoch 2004; Welti et al. 2003; Murdoch 2003). Therefore, the aims of this study were to evaluate the prevalence of atypical lower airway infections using nested PCR (nPCR) method in patients with various types of immunosuppression and to analyze clinical characteristics of these infections.
2 Methods
2.1 Patients
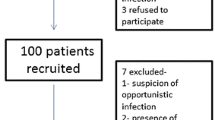
The study protocol was approved by an Institutional Bioethics Committee. The study group consisted of 80 non-HIV immunocompromised patients with different underlying diseases and clinical and radiological signs of pulmonary infection. Due to incomplete data on eight patients, 72 patients were eligible for final analysis (median age 58; range 16–79 years; F/M – 21/51). The patients were treated in a large multidisciplinary university hospital and in a specialized center for hematology and hematologic oncology in Warsaw, Poland. All met the following inclusion criteria: (1) known immunosuppression; (2) clinical or radiological signs and symptoms of pulmonary infection; and (3) signed informed consent for diagnostic bronchoscopy. Immunosuppression was defined as: (1) hematologic diseases or malignancies (HDM); or (2) immunosuppressive chemotherapy due to any malignant disease; or (3) immunosuppressive treatment due to solid organ or hematologic stem cell transplantation (SOTR); or (4) immunosuppressive therapy due to autoimmune or other diseases; or (5) miscellaneous chronic diseases that could affect the immune state (MISC group).
Clinical signs and symptoms suggestive of lower airway infection included recent cough, fever, dyspnea, or auscultatory findings. Radiological findings consistent with pulmonary infection were defined as the presence of the following pulmonary abnormalities: single or multifocal consolidations, areas of ground glass opacity, pulmonary nodules, interstitial pattern which could not have been explained by other causes, such as e.g. progression of lung tumors or new lung metastases. Exclusion criteria were the following: (1) known AIDS or positive result of HIV test; (2) contraindications to diagnostic bronchoscopy, i.e., unstable hemodynamic status, gas exchange abnormalities resulting in hypoxemia (SaO2 below 92 %) despite low flow oxygen therapy; and (3) respiratory failure requiring mechanical ventilation.
2.2 Bronchoscopy Procedure
All patients underwent fiberoptic bronchoscopy under local anesthesia. The insertion of a bronchoscope (Olympus BF 1 T180 or Pentax EB 1970 K; Tokyo, Japan) was preceded by premedication with atropine sulphate 0.5 mg s.c. and midazolam 7.5 mg p.o., and by local anesthesia of the upper airways with 2 % lidocaine. Suction was avoided in the upper airways and trachea to minimize contamination of the working channel of the bronchoscope. Additional portions of lidocaine were applied to the lower airways when necessary. After visual inspection of the lower airways, bronchoscope was wedged in segmental or sub-segmental bronchus in accordance with the localization of radiological abnormalities. In case of no relevant radiological abnormalities, bronchoscope was wedged in the medial or lateral segment of the right middle lobe (RB4 or RB5). Two hundred milliliters of sterile, pre-warmed (37 °C) 0.9 % saline solution were instilled either in ten 20 ml portions or four 50 ml portions and withdrawn by gentle suction. Bronchoalveolar lavage fluid (BALF) was collected in sterile polypropylene tubes.
2.3 Microbiological Procedure
Samples of BALF were sent for microbiological examination including direct microscopy, cultures, and fungal antigen detection, when appropriate. One milliliter samples of BALF were frozen at −20 °C. Total DNA was extracted from 200 μl of BALF, using EXTRAcell® isolation kit. Commercial qualitative amplification assay (PNEUMOTRIS oligomix Alert Kit®), based on nested PCR method, was used to detect specific DNA sequences of L. pneumophila, C. pneumoniae, and M. pneumoniae in defrozen BALF samples. Also BETA-GLOBIN oligomix Alert Kit®, which uses the human β-globin gene as a standard, was used as an external control of DNA extraction and amplification. All reagents described above were supplied by Nanogen Advanced Diagnostics S.r.L. (Turin, Italy), and all investigations were performed in accordance to the manufacturer’s instructions. A presumed limit of detection (LOD) of the PCR assay used was established as a few dozen copies/ml.
2.4 Data Collection and Analysis
Data on clinical and radiological signs and symptoms, and the results of microbiological examination of BALF were retrospectively collected and loaded in an electronic database. Additionally, results of other microbiological studies, including blood samples, throat swabs, sputum, urine, or stool were also analyzed.
Consistently with the aim of the study, results were assessed in patients with different types of immunosuppression.
Quantitative variables were presented as median, interquartile range (IQR) and/or ranges, while qualitative variables were presented as number and percentage. A non-parametric Mann-Whitney U test or Chi-squared test was used to assess the difference between variables in different groups. A p-value below 0.05 was considered statistically significant. Statistical analysis was performed using a statistical software package (STATISTICA, ver. 9.0, StatSoft Inc., Tulsa, OK).
3 Results
Demographics and data on the underlying diseases are presented in Table 1. Patients were unevenly distributed, with 61 (84.7 %) in the HDM group, 8 (11.1 %) in the MISC group, and 3 (4.2 %) patients in the SORT group. The most common underlying disease was acute myeloid leukemia (AML) which was responsible for almost one third of all causes of immunosuppression. AML was followed by chronic lymphocytic leukemia (n = 10; 13.9 % of causes) and non-Hodgkin lymphoma (n = 9; 12.5 % of causes).
Clinical signs and symptoms as well as radiographic data are demonstrated in Table 2. The major clinical symptoms were fever found in 54 (75.0 %) patients and cough reported by 30 (41.6 %) patients. There were no typical signs and symptoms of lower airway infection in 9 (12.5 %) patients, and pulmonary disease in these patients was diagnosed based on the new radiological findings. Chest radiographs and thorax CT scans were available in 71 (98.6 %) and 66 (91.7 %) of patients, respectively. The most common radiographic manifestation was lung consolidation, found in 50 (69.4 %) patients. There was a predominance of bilateral radiographic lung involvement, which was demonstrated in about half of patients, i.e., in 37/71 (52.1 %) and 46/66 (69.7 %) patients as based on chest radiograph and thorax CT scan, respectively. Isolated right lung involvement was found in 23 chest radiographs and 13 thorax CT scans.
Table 3 presents the clinical, radiological and microbiological characteristics of 9 patients in whom DNA of atypical pathogens was identified in BALF. In none of 72 samples specific L. pneumophila DNA sequences were found. M. pneumoniae specific DNA was identified in samples collected from 7 (9.7 %) patients. Two samples (2.8 %) tested positively for C. pneumoniae DNA. In all patients with identified atypical pathogens, fever was the most commonly reported symptom. In 6 out of the 9 patients bilateral lung involvement was demonstrated. In 8 patients, co-infections with different microorganisms were detected based on BALF or blood microbiological studies. Despite broad spectrum antibiotic and antifungal therapy, 3 patients died. All those patients had positive results of blood culture, with A. baumanii and P. aeruginosa found in two and one patients, respectively.
4 Discussion
The present study demonstrates a low prevalence of atypical pulmonary infections in non-HIV immunocompromised patients. M. pneumoniae, C. pneumoniae and L. pneumophila were found in 9.7 %, 2.8 %, and 0 % of patients, respectively. Thus, the prevalence of these infections in this study was somewhat lower than that usually reported in immunocompetent patients with CAP (Capelastegui et al. 2012; Masiá et al. 2007). On the other hand, the percentage of patients in whom atypical pathogens (except L. pneumophila) were identified was slightly higher as compared with other studies in immunocompromised hosts (Cervera et al. 2006; Hohenthal et al. 2005; Jain et al. 2004; Danés et al. 2002). This difference can be easily explained by multiple factors that can influence the results of various studies. These include: environmental factors (community or hospital acquired infection), seasonal and local epidemiological situation, type, severity and duration of immunosuppression, methods applied for pathogen detection and identification, reporting method (per entire study group or per subgroup with specific cause of immunoincompetence), and treatment applied prior to microbiological sampling. Despite all these conditions, most authors agree that atypical pulmonary infections in immunocompromised hosts are rather uncommon. Depending on the source of data, typical bacteria, fungi, and viruses have been responsible for 18–51 %, 8–38 %, and 9–23 % of pulmonary infections in non-HIV immunocompromised patients, respectively (Camps Serra et al. 2008; Jain et al. 2004; Danés et al. 2002; Rañó et al. 2001). In addition, polymicrobial infections caused by the pathogens outlined above have been diagnosed in 7–13 % of patients. Atypical pathogens have been found in single cases only.
In this study, diagnosis of pulmonary infection caused by atypical bacteria was based on a sole microbiological test, i.e., identification of specific DNA sequences in lavage fluid collected directly from the site of infection. The role of fiberoptic bronchoscopy and bronchoalveolar lavage as a diagnostic tool in immunocompromised patients with pulmonary infiltrates is well established. It has been shown that an early bronchoscopy (<5 days) has a significantly higher diagnostic yield for pulmonary infections than the late bronchoscopy (78 vs. 23 %; p = 0.02) (Lucena et al. 2014). The role of diagnostic methods other than culture in the work-up of immunocompromised patients with pulmonary infections has also been positively verified, albeit ELISA tests for the detection of C. pneumoniae and/or M. pneumoniae antibodies have some limitations, due to well-known cross reactions with other Chlamydia and Mycoplasma species. Hohenthal et al. (2005) have shown that the use of PCR and antigen detection to identify infectious agents in BALF from patients with hematological malignancies significantly improves the diagnostic yield. Unfortunately, although M. pneumoniae and C. pneumoniae PCR tests were performed in 37 and 29 BALF samples, respectively, the authors have neither presented nor discussed these results. Similar to the present study, none of the BALF samples evaluated by Hohenthal et al. (2005) tested positively for Legionella spp. in PCR tests. There are, however, two points which should be mentioned when comparing the results of these two studies. Firstly, the number of BALF samples evaluated by Hohenthal et al. (2005) has been almost two-fold higher than that in the present study. Secondly, In the Finnish study both PCR method and cultures have been applied and there was one patient with a positive culture but negative Legionella spp. PCR test. Thus, we cannot exclude that some patients with legionellosis could have been found in the present study, had other than PCR diagnostic methods been used. Nevertheless, the results of both studies point to a very low prevalence of L. pneumophila pulmonary infection in immunocompromised patients. That seems inconsistent with the results of some earlier studies which showed that hematological malignancies are a significant risk factor (rate ratio 22.4) for L. pneumophila pneumonia (Marston et al. 1994). Furthermore, as the course of pulmonary infections in immunocompromised patients is often severe and L. pneumophila is a well-known pathogen responsible for severe pneumonias, a higher prevalence of this infection could be expected in immunocompromised patients. Therefore, some methodological issues that could have negatively influenced the prevalence of L. pneumophila infections found in the present study should be considered. The hypothesis that extremely low prevalence of L. pneumophila infection was related to false negative PCR results is highly unlikely. Contrary to the above mentioned data (positive L. pneumophila culture and false negative PCR test) numerous other studies demonstrate that Legionella PCR has a sensitivity equal to, or greater than, culture. A PCR test can give false negative results when polymerase inhibitors are present in the biological sample (Hammerschlag 2000). It has been shown that in M. pneumoniae infections, throat swabs are preferred over nasopharyngeal samples due to a lower rate of PCR inhibitors (Murdoch 2003). As PCR inhibitors are usually nonspecific, their presence would have caused false negative results not only in terms of L. pneumophila infection but also other pathogens, i.e., M. pneumoniae and C. pneumoniae. This was not the case in our study, as an external control of DNA extraction and amplification was used simultaneously and no inhibition was observed during this study. Early and adequate antibiotic therapy before sample collection can be another cause of false negative results of microbiological studies. In fact, a significant proportion of our patients (65.3 %), including 7/9 patients with atypical bacterial infection, had been treated with macrolides or fluoroquinolones before or at the time of diagnostic bronchoscopy. Prior studies in patients with pneumonia have shown that bronchoalveolar lavage performed within 3 days of antibiotic therapy onset has a diagnostic yield of 63.4 %, while the diagnostic value decreases to 57.6 % and 34.4 %, when lavage is done later on, before and after 14 days of treatment initiation, respectively (Kottmann et al. 2011). The argument against the confounding role of prior treatment for the results obtained in the present study is that PCR tests allow detecting genetic material of causative pathogen even a few weeks after initiation of antibiotic therapy (Welti et al. 2003).
Interestingly, atypical pathogens were identified in the present study exclusively in males. This may be partially explained by a higher proportion of males (71 %). Nevertheless, we believe this is not a sufficient explanation for this finding. Some gender-related differences in the incidence of atypical bacterial infections have also been reported in previous studies. Gutiérrez et al. (2006) have found the incidence of CAP caused by C. pneumoniae and L. pneumophila in the general population two-fold and ten-fold higher in males than in females, respectively. Age-related differences in the prevalence of atypical pathogen infections should also be mentioned. In the present study, median age of patients with M. pneumoniae infection was 51 years. This is somewhat inconsistent with Gutiérrez et al.’s (2006) findings who have reported the highest incidence of M.pneumoniae CAP in young and very elderly people, and the lowest between 45 and 64 years of age. To our knowledge, no specific data have been published on the gender-related or age-related differences in the prevalence of atypical pathogen infection in immunocompromised patients. Therefore, we could not confront our observation with any other. We realize that the number of patients with atypical pathogen infections is too small to draw unequivocal conclusions on the relationship between age or gender and the prevalence of M. pneumoniae and C. pneumoniae infections.
The mortality rate in our nine patients with M. pneumoniae or C. pneumoniae infection was relatively high (33.3 %), but we believe that neither was the course of disease nor mortality rate related exclusively to atypical bacterial infection. In this context, it should be underlined that in eight of these patients co-infection with other microorganisms was found (positive BALF and/or blood cultures). Systemic bacterial co-infection was proved in all three patients who died (A. baumanii and P. aeruginosa cultured from blood samples). This finding is consistent with the results of three earlier studies that have reported co-infection with at least one another pathogen in 33–64 %, 48–74 %, and 54–63 % patients with M. pneumoniae, C. pneumoniae, and L. pneumophila infections, respectively (Welti et al 2003; Gleason 2002; Hammerschlag 2000). Perhaps, destruction of the airway epithelial layer and ciliostatic effect of these pathogens, facilitate other bacterial infections.
We are aware of several limitations of this study. Due to a small sample size, 95 % confidence interval could be calculated as 2.9–17.0 % and 0.0–6.8 % for a proportion of M. pneumoniae and C. pneumoniae infections, respectively. These values may question the confidence of a low prevalence of atypical pathogen infection in the study group. There is a marked disproportion between the number of patients with different causes of immunosuppression. In fact, our study group included mainly patients with hematological malignancies; hence the results refer mostly to this group of immunocompromised patients. That is also why we could not analyze the relationship between underlying diseases and the prevalence or clinical course of atypical infections.
A significant limitation of our study is associated with the use of PCR only to identify atypical bacteria infection. In consequence, we were unable to assess and discuss potential false positive and false negative results. Previous studies, including that by Pignanelli et al. (2009), have shown that a concomitant use of two or more different tests provides a higher diagnostic accuracy. Thus, the question on the true etiology of lower respiratory tract infection in some of our patients is still pending. In cases in which we did not find any putative etiological agent, it could have been any of the common respiratory viruses (metapneumovirus, coronavirus, or bocavirus) that are not routinely detected. Therefore, use of wide-range diagnostic tool, e.g., FilmArray® Respiratory Panel based on multiplex nested PCR assay, could be helpful to improve outcome in immunocompromised patients (Dzieciatkowski et al. 2013).
In conclusion, we found that atypical lower airway infections are uncommon in immunocompromised patients. This particularly refers to L. pneumophila pneumonia. The majority of atypical pulmonary infections are co-infections rather than single pathogen infections.
References
Bartlett JG (2008) Is activity against “atypical” pathogens necessary in the treatment protocols for community-acquired pneumonia? Issues with combination therapy. Clin Infect Dis 47:S232–S236
Bonatti H, Pruett TL, Brandacher G, Hagspiel KD, Housseini AM, Sifri CD, Sawyer RG (2009) Pneumonia in solid organ recipients: spectrum of pathogens in 217 episodes. Transplant Proc 41:371–374
Camps Serra M, Cervera C, Pumarola T, Rañó A, Benito N, Torres A, Jiménez de Anta MT, Marcos MA (2008) Virological diagnosis in community-acquired pneumonia in immunocompromised patients. Eur Respir J 31:618–624
Capelastegui A, España PP, Bilbao A, Gamazo J, Medel F, Salgado J, Gorostiaga I, de Goicoechea MJ L, Gorordo I, Esteban C, Altube L, Quintana JM, Poblational Study of Pneumonia (PSoP) Group (2012) Etiology of community-acquired pneumonia in a population-based study: link between etiology and patients characteristics, process-of-care, clinical evolution and outcomes. BMC Infect Dis 12:134
Cervera C, Agustí C, Angeles Marcos M, Pumarola T, Cofán F, Navasa M, Pérez-Villa F, Torres A, Moreno A (2006) Microbiologic features and outcome of pneumonia in transplanted patients. Diagn Microbiol Infect Dis 55:47–54
Corti M, Palmero D, Eiguchi K (2009) Respiratory infections in immunocompromised patients. Curr Opin Pulm Med 15:209–217
Danés C, González-Martín J, Pumarola T, Rañó A, Benito N, Torres A, Moreno A, Rovira M, Puig de la Bellacasa J (2002) Pulmonary infiltrates in immunosuppressed patients: analysis of a diagnostic protocol. J Clin Microbiol 40:2134–2140
Di Stefano F, Verna N, Di Gioacchino M (2007) Cavitary Legionella pneumonia in a patient with immunodeficiency due to hyper-IgE syndrome. J Infect 54:e121–e123
Dzieciatkowski T, Przybylski M, Sulowska A, Rynans S, Mlynarczyk G, Swoboda-Kopec E (2013) Application of FilmArray assay for detection of respiratory tract infections in immunocompromised persons. Med Dosw Mikrobiol 65:181–185
Gleason PP (2002) The emerging role of atypical pathogens in community-acquired pneumonia. Pharmacotherapy 22:2S–11S, discussion 30S–32S
Gutiérrez F, Masiá M, Mirete C, Soldán B, Rodríguez JC, Padilla S, Hernández I, Royo G, Martin-Hidalgo A (2006) The influence of age and gender on the population-based incidence of community-acquired pneumonia caused by different microbial pathogens. J Infect 53:166–174
Hammerschlag MR (2000) Chlamydia pneumoniae and the lung. Eur Respir J 16:1001–1007
Heinemann M, Kern WV, Bunjes D, Marre R, Essig A (2000) Severe Chlamydia pneumoniae infection in patients with neutropenia: case reports and literature review. Clin Infect Dis 31:181–184
Hohenthal U, Itälä M, Salonen J, Sipilä J, Rantakokko-Jalava K, Meurman O, Nikoskelainen J, Vainionpää R, Kotilainen P (2005) Bronchoalveolar lavage in immunocompromised patients with haematological malignancy-value of new microbiological methods. Eur J Haematol 74:203–211
Jain P, Sandur S, Meli Y, Arroliga AC, Stoller JK, Mehta AC (2004) Role of flexible bronchoscopy in immunocompromised patients with lung infiltrates. Chest 125:712–722
Kottmann RM, Kelly J, Lyda E, Gurell M, Stalica J, Ormsby W, Moon K, Trawick D, Sime PJ (2011) Bronchoscopy with bronchoalveolar lavage: determinants of yield and impact on management in immunosuppressed patients. Thorax 66:823
Lucena CM, Torres A, Rovira M, Marcos MA, de la Bellacasa JP, Sánchez M, Domingo R, Gabarrus A, Mensa J, Agustí C (2014) Pulmonary complications in hematopoietic SCT: a prospective study. Bone Marrow Transplant 49:1293–1299
Marston BJ, Lipman HB, Breiman RF (1994) Surveillance for Legionnaires’ disease. Risk factors for morbidity and mortality. Arch Intern Med 154:2417–2422
Masiá M, Gutiérrez F, Padilla S, Soldán B, Mirete C, Shum C, Hernández I, Royo G, Martin-Hidalgo A (2007) Clinical characterisation of pneumonia caused by atypical pathogens combining classic and novel predictors. Clin Microbiol Infect 13:153–161
Murdoch DR (2003) Nucleic acid amplification tests for the diagnosis of pneumonia. Clin Infect Dis 36:1162–1170
Murdoch DR (2004) Molecular genetic methods in the diagnosis of lower respiratory tract infections. APMIS 112:713–727
Perez CR, Leigh MW (1991) Mycoplasma pneumoniae as the causative agent for pneumonia in the immunocompromised host. Chest 100:860–861
Pignanelli S, Shurdhi A, Delucca F, Donati M (2009) Simultaneous use of direct and indirect diagnostic techniques in atypical respiratory infections from Chlamydophila pneumoniae and Mycoplasma pneumoniae. J Clin Lab Anal 23:206–209
Rañó A, Agustí C, Jimenez P, Angrill J, Benito N, Danés C, González J, Rovira M, Pumarola T, Moreno A, Torres A (2001) Pulmonary infiltrates in non-HIV immunocompromised patients: a diagnostic approach using non-invasive and bronchoscopic procedures. Thorax 56:379–387
Rañó A, Agustí C, Benito N, Rovira M, Angrill J, Pumarola T, Torres A (2002) Prognostic factors of non-HIV immunocompromised patients with pulmonary infiltrates. Chest 122:253–261
Singanayagam A, Chalmers JD, Welte T (2014) Epidemiology of CAP in Europe. Eur Respir Monog 63:1–12
Sousa D, Justo I, Domínguez A, Manzur A, Izquierdo C, Ruiz L, Nebot M, Bayas JM, Celorrio JM, Varona W, Llinares P, Miguez E, Sánchez E, Carratalá J (2013) Community-acquired pneumonia in immunocompromised older patients: incidence, causative organisms and outcome. Clin Microbiol Infect 19:187–192
Welti M, Jaton K, Altwegg M, Sahli R, Wenger A, Bille J (2003) Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn Microbiol Infect Dis 45:85–95
Yu VL, Stout JE (2008) Community-acquired legionnaires disease: implications for underdiagnosis and laboratory testing. Clin Infect Dis 46:1365–1367
Acknowledgment
The authors gratefully acknowledge Warsaw Medical University and the Foundation for Patients with Hematological Diseases in Warsaw, Poland for the financial support that enabled the realization of the project.
Conflicts of Interest
The authors declare no conflicts of interst in relation to this article.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Grabczak, E.M., Krenke, R., Przybylski, M., Kolkowska-Lesniak, A., Chazan, R., Dzieciatkowski, T. (2016). Prevalence of Pulmonary Infections Caused by Atypical Pathogens in non-HIV Immunocompromised Patients. In: Pokorski, M. (eds) Pulmonary Infection and Inflammation. Advances in Experimental Medicine and Biology(), vol 935. Springer, Cham. https://doi.org/10.1007/5584_2016_28
Download citation
DOI: https://doi.org/10.1007/5584_2016_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-44484-0
Online ISBN: 978-3-319-44485-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)