Abstract
Poland has implemented the influenza surveillance system called Sentinel as of 2004. The system consists of both epidemiological and virological surveillance. It is an important tool for monitoring the situation in the entire country, coordinated by the National Influenza Center situated at the National Institute of Public Health-National Institute of Hygiene (NIPH-NIH) in Warsaw, Poland. In the 2015/2016 epidemic season, more than 1600 samples were tested in the Sentinel System, of which 38.6% were positive. The samples were evaluated in seven age-groups: 0–4, 5–9, 10–14, 15–25, 26–44, 45–64, and over 65 years of age. Significant differences were reported in the number of confirmed cases of infection caused by influenza and influenza-like viruses, depending on the age-group. The highest number of confirmed cases of infections was reported for the age range of 26–44 years, accounting for 30% of the total. In each age-group, the presence of infection caused by influenza-like viruses, collectively accounting for only 3.8% of all positive tests, was also confirmed. Weekly reports generated by the Sentinel System enable to determine and control a current influenza activity in the country, which is of essential importance in case of the emergence of a new strain with a pandemic potential.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Over decades, the world has been attacked by many pandemics. The greatest havoc in the twentieth century was brought about by Spanish influenza, caused by the virus subtype A/H1N1/, which took place in the years 1918/1919. According to the latest data, it caused about 50 million deaths and large economic losses (WHO 2004). For this reason, the WHO launched an international influenza surveillance system in 1947. It served to observe changes in the structure of the influenza virus and to rapidly detect newly formed virus variants (Życińska and Brydak 2007). In 2011, the network of surveillance has evolved to be coordinated by the WHO Global Influenza Surveillance and Response System (GISRS) (Bednarska et al. 2016a, b).
The best proof of the importance of the preparation of virological and epidemiological reports are those drawn up in 1997 in the National Influenza Center in Hong Kong after the emergence of the avian influenza virus A/H5N1/ (Highly Pathogenic Avian Influenza – HPAI) (Brydak 2008). Thanks to the reports it was possible to react quickly and focus on preventing the spread of the virus. A quick reaction was essential because according to the latest WHO estimates, 60% of infections caused by this subtype are fatal (Brydak 2014). In 2003, the National Center of Influenza in Rotterdam identified a different dangerous subtype of the HPAI determined as A/H7N7/ (Elbers et al. 2004), which reaffirms the notion of the influenza virus constantly undergoing modifications. In view of this unpredictability and the virus spread in the world, National Influenza Centers have been created in many countries. By 2008, 122 such centers were in operation throughout the world (Brydak 2008). Currently, there are 142 National Influenza Centers that act as reference centers in different countries. In Poland, this center is placed at the National Institute of Public Health-National Institute of Hygiene (NIPH-NIH). Apart from the GISRS, surveillance of influenza activity in Europe also is conducted by the European Influenza Surveillance Network (EISN), which is overseen by the European Center for Disease Control and Prevention (ECDC). The EISN conducts both epidemiological and virological surveillance. Data on the situation in European countries are gathered in the platform of the European Surveillance System (TESS) that issues weekly reports assessing the current status of influenza spread in Europe (Bednarska et al. 2016a, b).
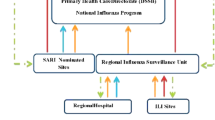
The Sentinel System of epidemiological and virological surveillance is in place in Poland. The initiation of the system was made possible after fulfilling the international requirements imposed by the European Influenza Surveillance Scheme (EISS) (Brydak 2008). Sentinel works in cooperation with general practitioners and 16 Voivodeship (province) Sanitary Epidemiological Stations (VSES) present. This enables a continuous observation of virological and epidemiological situation in the country (Romanowska and Brydak 2007). Family doctors, who make up at least 1% of all doctors, are obliged to collect clinical samples from patients and to report on the epidemiological situation in a given area (Woźniak-Kosek and Brydak 2013). The samples are then sent to the VSES where diagnostic tests are performed using biomolecular methods to confirm the presence of influenza and influenza-like viruses.
As of the 2013/2014 epidemic season, influenza surveillance has been modified by expanding the age-group stratification for recording infections with influenza and influenza-like viruses, which improves the clarity of results (Bednarska et al. 2016a, b). The results are sent weekly by representatives of VSES, enabling to report by the National Influenza Center to the GISRS and EISN networks. From the data collected, epidemiological reports are generated, posted every week on the website of the National Institute of Public Health-National Institute of Hygiene. The National Influenza Center, acting as a reference place, is responsible for controlling the performance of VSES by random checks to confirm the positive results of samples obtained from the VSES.
The goal of the present study was to investigate the efficacy of epidemiological data elaboration in the Sentinel System with regard to the 2015/2016 influenza epidemic season in Poland.
2 Methods
2.1 Material
The study was approved by an institutional Ethics Committee and it was conducted in accordance with the Declaration of Helsinki for Human Research. In the 2015/2016 epidemic season, 1625 samples acquired by the country 16 VSES were tested in the Sentinel System. The clinical material was derived from swabs taken from the nose and throat. The results were evaluated in the following age-groups: 0–4, 5–9, 10–14, 15–25, 26–44, 45–64, and >65 years of age.
2.2 Isolation of RNA
RNA isolation was carried out using the Maxwell 16 Viral Total Nucleic Acid Purification Kit (Promega Corporation; Madison, WI) according to the manufacturer’s instructions. The samples were suspended in physiological saline. The final volume of the sample after elution was 50 μl.
2.3 Real-Time Reverse Transcription Polymerase Chain Reaction (rRT-PCR)
The real-time reverse transcription (rRT) polymerase chain reaction (PCR) was performed using a Light Thermocycler 2.0 System (Roche Diagnostics; Rotkreuz, Switzerland). The final reaction volume in the capillaries was 20 μl; 5 μl of which was the previously isolated RNA. Primers and probes necessary to carry out the reaction were obtained through the Influenza Reagent Resource (IRR) program from the US Center for Disease Control (CDC). The reaction mixture consisted of bovine serum albumin (BSA) buffer, MgSO4, RNase free water, and a SuperScript III/Platinum Taq Mix (Invitrogen Life Technologies-Thermo Fisher Scientific; Carlsbad, CA). Viruses included in the vaccine for the 2015/2016 epidemic season were the following: A/California/7/2009(H1N1)pdm09 and A/Switzerland/9715293/2013, and B/Phuket/3073/2013 functioned as a positive control. The negative control was RNase free water provided in the kit. Before the amplification process, in order to obtain cDNA, the RNA was subjected to reverse transcription (30 min 50 °C). The resulting DNA was subjected to a process of initiation (1 cycle 95 °C for 2 min), followed by 45 cycles of amplification: denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 20 s.
2.4 Conventional Multiplex RT-PCR
To confirm the genetic material of influenza-like viruses, RT-PCR reaction using an RV12 ACE Detection Kit (Seegene; Seul, South Korea) was performed. Influenza A and B viruses, human adenovirus, human respiratory syncytial A and B viruses, human metapneumovirus, human OC43 and 229E/NL63 coronaviruses, human parainfluenza 1, 2, and 3 viruses, and human rhinovirus A/B could be detected. Random hexamer-primed cDNA synthesis products were generated using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific; Carlsbad, CA) according to the manufacturer’s instructions. Each cDNA preparation was subjected to the RV12 PCR procedure. Finally, amplicons were detected by gel electrophoresis.
3 Results
In the 2015/2016 epidemic season, 1625 tests were run in the Sentinel System, out of which 627 (38.6%) samples confirmed the presence of influenza and influenza-like viruses (Table 1). Infections caused only by influenza virus were present in 603 cases (37.1%). Influenza virus type A and B were the equally predominant contagions, accounting for 50.6% and 49.4% of infections, respectively. Concerning the influenza A virus, the predominant subtype was A/H1N1/pdm09, which accounted for 44.1% of all infections. There was only one case (0.2%) of the presence of subtype A/H3N2/, and 6.3% of influenza A cases remained unsubtyped.
Positive samples were evaluated in the seven age-group division. The majority of confirmed influenza viruses were observed in the group of persons aged 26–44 – 181 (30.0%) cases and in children aged 5–9 – 145 (24.1%) cases, and the least in those aged over 65 (4.0%) (Table 1). Influenza virus type B predominated in the youngest groups of 0–4, 5–9, and 10–14 years of age, as opposed to subtype A/H1N1/pdm09 that predominated in the other age-groups (Table 2).
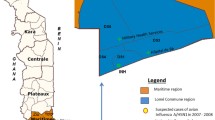
Most samples were tested in the province of Western Pomerania (n = 390) and the fewest in Malopolska (n = 12). Most confirmations of infection with influenza virus type A and subtype A/H1N1/pdm09 were recorded in the province of Podkarpackie, while the influenza virus type B was found most frequently in Western Pomerania (Table 3).
Influenza-like virus infections were confirmed only in 24 samples, which accounted for 1.5% of all those tested. The dominant influenza-like virus was RSV (41.7%). Moreover, the presence of the genetic material of viruses such as PIV3 (29.2%), adenovirus (12.5%) PIV1 (8.3%), PIV2 (4.2%), and human coronavirus (4.2%) was confirmed. The largest percentage of influenza-like viruses was reported in the group of persons aged 26–44 (29.2% of cases), while in those aged 10–14 and over 65 no influenza-like virus infections were reported (Table 2).
There were three co-infections detected in individuals of 7 (influenza type A and B), 12 (influenza A/H1N1/pdm09 and B), and 67 years of age (influenza A/H1N1/pdm09 and B).
4 Discussion
The Sentinel System is an important part of virological and epidemiological surveillance. Thanks to the data reported from VSES to the National Influenza Center of the National Institute of Public Health-National Institute of Hygiene (NIPH-NIH 2016), it is possible to determine the type of influenza and influenza-like viruses currently circulating in the population. That is essential for the proper selection of vaccine strains for the epidemic season and also provides information on the emergence of new viral subtypes and on antigenic changes in the circulating strains; all of which helps prevent the development of a pandemic.
Primary care doctors are the foremost participants of epidemiological surveillance. They are obliged to record cases of infections. The number of participating doctors ranges from 1 to 5% of all country physicians each season. The 2015/2016 season considered in the present study involved 524 primary care physicians; the number was comparable with those in previous seasons (Table 4). It is worth emphasizing that doctors work pro publico bono as they do not receive compensation for participation in surveillance.
The Sentinel System confirmed 627 cases of infections caused by influenza viruses in the 2015/2016 epidemic season. This figure is almost 2.7-fold greater than the 2014/2015 result and 3.5-fold greater than 2013/2014 result (Table 4). That speaks for an extremely good and continuously increasing efficiency of the surveillance system in Poland, with passing years.
In the 2015/2016 epidemic season, same as in the preceding one, an equal predomination of influenza type A and B viruses was observed. However, subtypes of the influenza A virus differed, with A/H1N1/pdm09 predominating in 2015/2016 (16.4%) and A/H3N2/ (22.6%) in 2014/2015 (Bednarska et al. 2016a, b; WHO 2015). The results noted in Poland correspond well with those in other European countries, for instance, in Austria or Finland where the A/H1N1/pdm09 subtype also predominated (FluNews Europe 2016).
The present findings also underscore the possibility of regional variability in the influenza virus subtype. Type B virus predominated in western Poland whereas type A/H1N1/pdm09 did in eastern Poland (Table 3). This situation also was reflected in the results obtained in neighboring countries. On the western side, in Germany, type B virus predominated, whereas on the eastern side, in Ukraine, subtype A/H1N1/pdm09 predominated (FluNews Europe 2016), which is explicable by a readily spread of the virus across borders.
References
Bednarska K, Hallmann-Szelińska E, Kondratiuk K, Rabczenko D, Brydak LB (2016a) Novelties in influenza surveillance in Poland. Probl Hig Epidemiol (2):101–105 (Article in Polish)
Bednarska K, Hallmann-Szelińska E, Kondratiuk K, Brydak LB (2016b) Influenza surveillance. Postępy Hig Med Dosw 70:313–318 (Article in Polish)
Brydak LB (2008) Influenza, pandemic flu, myth or real threat? Rythm, Warsaw, 1–492 (Article in Polish)
Brydak LB (2014) Influenza – the greatest master of metamorphosis – constant puzzle. JHPOR 2:4–11
Elbers ARW, Fabri THF, de Vries TS, de Wit JJ, Pijpers A (2004) Koch G (2004) The highly pathogenic avian influenza A (H7N7) virus epidemic in the Netherlands in 2003 – lessons learned from the first five outbreaks. Avian Diseases: September 48(3):691–705
FluNews Europe (2016) Available from: https://flunewseurope.org/. Accessed 22 Nov 2016
NIPH-NIH (2016) Influenza and influenza-like illness in Poland Available from: http://wwwold.pzh.gov.pl/oldpage/epimeld/grypa/index.htm. Accessed 22 Nov 2016
Romanowska M, Brydak LB (2007) The role of surveillance in the fight against influenza including pandemic. Fam Med Prim Care Rev 9(3): 823–829 (Article in Polish)
WHO (2004) Wkly Epidemiol Rec 79:25–40
WHO (2015) Review of the 2014–2015 influenza season in the northern hemisphere. Available from: http://www.who.int/wer/2015/wer9023/en/. Accessed 22 Nov 2016
Woźniak-Kosek A, Brydak LB (2013) The epidemiological and virological surveillance of influenza in the Polish population – Sentinel. Fam Med Prim Care Rev 15(3):483–485
Życińska K, Brydak LB (2007) Influenza and flu prophylaxis – a current medical issue. Pol Arch Med Wewn 117(10): 464–469 (Article in Polish)
Acknowledgments
This work was funded in part by the NIPH-NIH thematic grant 5/EM.1. The authors would like to acknowledge physicians and employees of VSESs participating in sentinel and non-sentinel programs for their input into the influenza surveillance in Poland.
Conflicts of Interest
The authors declare no conflicts of interest in relation to this article.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Cieślak, K., Kowalczyk, D., Szymański, K., Brydak, L.B. (2017). The Sentinel System as the Main Influenza Surveillance Tool. In: Pokorski, M. (eds) Respiratory System Diseases. Advances in Experimental Medicine and Biology(), vol 980. Springer, Cham. https://doi.org/10.1007/5584_2016_205
Download citation
DOI: https://doi.org/10.1007/5584_2016_205
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59497-2
Online ISBN: 978-3-319-59498-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)