Abstract
Phytophthora species are well-known as destructive plant pathogens, especially in natural ecosystems. It is ironic, therefore, how little is known regarding the Phytophthora diversity in South African natural woody ecosystems. In this study, Phytophthora species were isolated using standard baiting techniques from 182 soil and water samples and these were identified based on ITS and coxl sequence data. The 171 resulting Phytophthora isolates resided in 14 taxa including six known species (P. multivora, P. capensis, P. cryptogea, P. frigida, P. cinnamomi, P. cinnamomi var. parvispora), the known but as yet unnamed Phytophthora sp. PgChlamydo, P. sp. emzansi, and P sp. Kununurra and five novel taxa referred to as P. sp. stellaris, P. sp. Umtamvuna P. sp. canthium, P. sp. xWS, P. sp. xHennops. Four of the new taxa were found exclusively in water and two of these are hybrids. The most commonly isolated species from soil was P. multivora, a species recently described from Western Australia. Phytophthora frigida was isolated for the first time from stream water. With the exception of P. cinnamomi, very little is known regarding the biology, epidemiology or origin of Phytophthora in South Africa.
Similar content being viewed by others
Introduction
Less than 2% of the land surface of South Africa is covered with indigenous forests. The larger part of the country is grassland and dry savanna woodland such as semi-dessert with small shrubs and Acacia trees (Grundy & Wynberg 2001). Savanna woodlands cover 35–40%, while plantation forests cover 1.5% of the land area. Before 1940, small privately-owned plantations of Acacia or Eucalyptus species in Western Cape were associated with agriculture to protect crops from wind erosion and subsequent sand drift. After 1945, the Department of Forestry was established to protect indigenous forest by establishing a plantation industry based on non-native species such as Eucalyptus spp. from Australia and Pinus spp. from north and central America (Burgess & Wingfield 2001, Grundy & Wynberg 2001). South Africa is also home to three of the world’s 25 Biodiversity host-spots (https://doi.org/www.biodiversityhotspots.org) and many studies have been conducted to document and conserve animal and plant biodiversity. In contrast, there has been relatively little work on fungal biodiversity (Crous et al. 2006, Marincowitz et al. 2008) and almost nothing is known regarding the endemic oomycetes, which are broadly treated with the fungi.
Several common Phytophthora species have been recovered from agricultural landscapes in South Africa, notably P. cinnamomi, P. cactorum, P. citrophthora, P. citricola, P. megasperma, P. cryptogea, P. drechsleri, P. infestans, P. nicotianae, P. syringae, and P. porri (Crous et al. 2000). Eight Phytophthora spp. have been recovered from plantations of non-native species, mainly located in KwaZulu-Natal, Eastern and Western Cape, and Mpumalanga. These include P. boehmeriae, P. cinnamomi, P. cryptogea, P. nicotianae, P. meadii, P. frigida, and P. alticola (Zeijlemaker 1971, Bumbieris 1976, Wingfield & Knox-Davies 1980, Linde et al. 1994, Roux & Wingfield 1997, Maseko et al. 2001, Maseko et al. 2007). All the Phytophthora species commonly found in agriculture and forestry are considered introductions to South Africa. Phytophthora alticola and P. frigida are the only species known exclusively from South Africa, and they could be endemic to the region.
The species most studied from natural ecosystems in South Africa is the devastating pathogen P. cinnamomi (von Broembsen 1984a, von Broembsen & Kruger 1985). It is also commonly recovered from dying Proteaceae, including commercially cultivated members such as Protea spp., Leucodendron spp., and Leucospermum spp., mostly in the Cape Province (Knox-Davies 1975, von Broembsen 1984b, von Broembsen & Kruger 1985).
In 2010, P. capensis and P. sp. emzansiFootnote 1 were identified from the cultivated endemic shrubs Agathosma betulina, olea capensis, and Curtisia dentate, and also stream water, in Cape Province (Bezuidenhout et al. 2010). The original isolates of P. capensis had been reported as “P. citricola complex” (CIT4) in an earlier study (oudemans et al. 1994). Additionally, P. cinnamomi, P. cinnamomi var. parvispora, P. citricola, P. cryptogea, P. dreschsleri, P. multivora, P. nicotianae, and P. plurivora were identified on diseased Agathosma spp. in commercial fields and nurseries (Bezuidenhout et al. 2010). In a recent study, P. sp. PgChlamydo and several hybrid species have recently been reported from a stream within a botanical garden in Gauteng (Nagel et al. 2013b). other than the latter two studies and the considerable body of literature on the impact of P. cinnamomi in natural and managed ecosystems in South Africa, there have been no recent studies examining the diversity, biology or impact of Phytophthora species in natural ecosystems. The aim of the present study was thus to broaden our knowledge of the genus in the country by collections of the genus from such environments.
Materials and Methods
Sampling and isolation
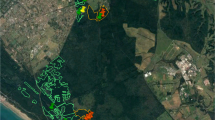
Sampling was conducted in five provinces of South Africa in different climatic zones (Table 1, Fig. 1): Mpumulunga (MP; sub-tropical climate), 26 soil samples and 3 water samples from natural vegetation (one soil sample was from a Eucalyptus plantation); Gauteng (GT; temperate climate, over 2000 m elevation), 4 water samples from a botanical garden; Western Cape (WC; Mediterranean climate), 6 soil samples and 2 water samples from natural vegetation; Eastern Cape, Umtamvuna Nature Reserve (UM; temperate climate), 21 soil samples and 6 water samples for filtering from natural vegetation; KwaZulu-Natal, Pietermaritzburg (PMB; temperate climate), 16 soil samples, 12 water samples for filtering and 13 water samples for baiting from a botanical garden; and KwaZulu-Natal, Ingeli Forest Reserve (ING; subtropical climate), 52 soil samples, 9 water samples for filtering and one water sample for baiting from natural forest.
For soil baiting, 2–300 g of soil was placed in a container (12 × 22 cm stainless steel, Sunnex, UK) containing 800 mL of non-sterile distilled water. Floating litter was removed, and two intact and edge-excised Rhododendron (R. indioum Claude Goyet) and pear leaves were floated on the surface of water for up to 7 d until lesions appeared. The margin of the necrotic regions was excised and cut into to small pieces (5 × 5 mm) and placed onto Phytophthora selective medium, NARPH (Hüberli et al. 2000). The plates were incubated at room temperature, of approximately 22 °C for 3–7 d, and mycelium on the plates was transferred to cornmeal agar (CMA).
Water samples (1 L) were baited in the laboratory using a technique modified from Jung et al. (1996), where stream water was placed in a 12 × 22 cm stainless steel container (Sunnex, UK), baited with Rhododendron indioum leaves and whole pear and apple fruits (previously washed and surface-sterilized with 95% ethanol). The leaves were collected after 3–4 d when necrotic symptoms were visible, and the fruits after 7 d. The baits were rinsed with sterilized dH2O and placed on paper towels to remove excess water, sterilized in 95% ethanol for 10–20 s, rinsed in sterilized dH2o and dried with paper towel. Sections containing lesions were then excised and plated onto NARPH medium and purified as described above. other water samples were filtered shortly after collection, through a 47 mm circle filter paper with 0.45 urn pore size (Whatman™, Kent, UK) using a filtering funnel and glass flask connected to a vacuum pump. Filters were then placed topside down onto the surface of NARPH medium for 2 d, after which they were removed and individual colonies were transferred to new plates.
Single hyphal tips of all putative Phytophthora spp. from the isolations using the various techniques were transferred to V8 agar. After purification, they were stored in the culture collection (CMW) of the Forestry and Agricultural Biotechnology Institute at the University of Pretoria.
Identification of Phytophthora species
Mycelium of isolates grown in 10% V8 broth was harvested, washed with sterile distilled water, removed of excess water with filter paper, placed in a 2 mL microfuge tubes, and lyophilised with VirTris Advantage BenchTop Tray Lyophilizer (SP Scientific, UK) overnight. The dried mycelium was then transferred to new microfuge tubes with two 3 mm metal beads. Extraction of total genomic DNA and amplification of target genes by polymerase chain reaction was carried out using a modification of the protocol described by Winton & Hansen (2001).
The Internal Transcribed Spacer regions of the rDNA (ITS1, 5.8S, and ITS2) were amplified using the primers ITS6 (Cooke & Duncan 1997) and ITS4 (White et al. 1990) and the cytochrome oxidase subunit I (coxI) was amplified using the primers FM84 and FM83 (Martin & Tooley 2003) with annealing temperatures of 55 °C and 50 °C respectively. Amplified DNA was purified with a high pure PCR product purification kit (Roche, RSA) and sequenced with the same primers. Cloning was carried out using pGEM-T® Easy vectors (Promega, USA) following manufacturer’s instructions when the sequences of the isolates could not be read due to DNA polymorphism.
Sequences of the isolates were uploaded and aligned in Geneious v. R6 (Biomatters; available from https://doi.org/www.geneious.com/). The most appropriate substitution model was determined using jModelTest (Posada 2008). The TIM3+G (ITS) and GTR+I+R (coxI) model were selected and used in the Bayesian analysis (Ronquist & Huelsenbeck 2003). All sequences for the isolates considered in this study were submitted to GenBank and given in Figs 2–3.
A phylogram based on ITS sequence data indicating the placement of the 14 Phytophthora species recovered in this study in relation to closely related taxa. Numbers in circles represent the Clade as designated by Cooke et al. (2000). Numbers above the branch represent the bootstrap support based on parsimony analysis.
Phylograms based on coxI sequence data indicating the placement of undescribed Phytophthora taxa recovered in this study in relation to closely related taxa in (A) Phytophthora Clade 6 and (B) Phytophthora Clade 9. Bootstrap support is given above the line. Numbers above the branch represent the bootstrap support based on parsimony analysis.
Results
Isolation
Isolation success varied between sites and the methods used (Table 1); soil samples from the Western Cape, Mpumulunga, Ingeli Forest Reserve, Umtamvuna and Pietermaritzburg yielded a success rate of 100, 28, 38, 65 and 81% respectively. Isolation success from water-baited samples in the Western Cape, Mpumulunga, Gauteng, Ingeli Forest Reserve, and Pietermaritzburg gave a success rate of 0, 50, 100, 12.5 and 53.8% respectively. Isolation by water filtering from Ingeli Forest Reserve, Umtamvuna and Pietermaritzburg gave success rates of 55%, 50% and 61% respectively. In total, 171 Phytophthora isolates were recovered; 98 from soil, 32 from water baiting and 41 from waterfiltering (Table 1).
Identification of Phytophthora isolates
ITS sequence data were obtained for all isolates, and their identity confirmed by firstly conducting BLAST searched in GenBank (https://doi.org/www.ncbi.nlm.nih.gov/genbank/) and secondly by alignment to sequences of type isolates from original publications or, if these were unavailable, to the representative isolates selected for the oomycete barcode paper of Robideau et al. (2011). A maximum of three sequences per taxa were selected for inclusion in the complete ITS phylogenetic analysis (TreeBASE 14082). There were three taxa for which the ITS could not be directly sequenced; one group residing in Clade 6 and two in Clade 9 of the phylogeny (Fig. 2). In each case, the ITS product of representative isolates from each group were cloned and 10 cloned fragments sequenced (11 isolates of P. sp. xWS, 8 isolates of P. sp. xHennops and 3 isolates of P. sp. stellaris). These were then aligned to known taxa and ITS alleles representing each of the taxa were selected for inclusion in the complete ITS phylogenetic analysis.
The aligned ITS dataset consisted of 945 characters of which 492 were parsimony informative. Analysis resulted in 24 trees of 1887 steps (CI = 0.54, RI = 0.91) (Fig. 2). Fourteen Phytophthora spp. were identified from amongst the 173 isolates (Table 1, Fig. 2). of these, six were of known species (P. multivora, P. capensis, P. frigida, P. cinnamomi, P. cinnamomi var. parvispora, and P. cryptogea), three species matched previously designated taxa (P. sp. emzansi, P. sp. PgChlamydo, P.sp. Kununurra) and five taxa did not correspond to any known species and are designated here as P. sp. xWS, P. sp. stellaris, P. sp. xHennops, P. sp. Umtamvuna and P. sp. canthium.
For three of the unknown taxa, P. sp. xWS, P. sp. xHennops and some isolates of P. sp. stellaris, direct sequencing of the ITS region failed. After cloning of ITS amplicons, each of these species yielded at least ITS alleles corresponding in the phylogenetic analysis to other taxa. For P. sp. xWS, alleles corresponding to P. thermophila and P. amnicola in Clade 6 were obtained (12 SNPs). For P. sp. xHennops, alleles corresponding to P. hydropathica and an unknown species in Clade 9 were obtained (24 SNPs). For P. sp. stellaris, direct sequencing was not possible due to a 1bp indel in one of the ITS alleles, however the remaining variation (3 bp) between the ITS alleles was considered to be within the range of intraspecific variation. P. sp. stellaris is most similar to P. insolata but differs by 36 bp (4.4%), P. sp Umtamvuna differs from P. hydropathica by 23 bp (2.55%) and from P. sp. Kununurra by 19 bp (3.05%) and P. sp. canthium differs from P. kernoviae by 42 bp (5.1%). In the phylogenetics analysis Clade 9, separates into four sub-clades (Fig. 2).
coxI sequence data were obtained for most isolates and those of P. multivora, P. capensis, P. cryptogea, P. frigida, P. cinnamomi, P. cinnamomi var. parvispora and P. sp. emzansi returned 100% matches to corresponding species in Blast searches on GenBank (data not shown). Phylogenic analyses was conducted including all related species in Clade 6 (Nagel et al. 2013), and those for which sequence data was available from Clade 9 (Fig. 3). All isolates designated as P. sp. stellaris had identical coxI alleles. Isolates designated as P. sp. xWS based on ITS sequence data had coxI alleles corresponding to either P. thermophila or P. amnicola. Isolates designated as P. sp. xHennops had coxI alleles corresponding to either P. sp. Kununurra or P. hydropathica. The latter two taxa, P. sp. xWS and P. sp. xHennops are considered to be hybrids and are designated as such by the use of the “x”.
Distribution of Phytophthora isolates
With the exception of three isolates of Phytophthora multivora recovered from dying Rapanea collected in the Harlod Porter Botanical Garden in Western Cape Province, all other isolates were recovered from soil associated with asymptomatic plants or from water (Table 2, Fig. 1). Phytophthora multivora was the most frequently isolated species (40% of all isolates) and was recovered from all locations except GT (Fig. 1). It was almost always recovered from the soil, except for three isolates recovered from filtered water (Table 2). P. cinnamomi (9.25% of the isolates) was also recovered only from soil in UM, PMB and MP (Table 2, Fig. 1). of the other known species, P. capensis (4.6% of the isolates) was recovered from soil at PMB, UM and ING and once from filtered water in ING, P. cryptogea was recovered once from soil in ING and P. cinnamomi var parvispora was recovered once from filtered water in UMT (Table 2, Fig. 1).
Of the previously designated taxa, P. sp. emzansi (2.9% of the isolates) was recovered only from Ingeli forest, where it was found in both soil and filtered water, P. sp. PgChlamydo (5.2% of the isolates) was recovered from ING, PMB and MP, predominantly from water, but the isolate from MP was from soil on a riverbank (Table 2, Fig. 1). An isolate with a 100% ITS sequence match to P. sp. Kununurra was recovered once from soil in MP.
The remaining isolations, with the exception of the isolate designated as P. sp. canthium from Umtamvuna, were of previously undescribed taxa recovered from water either through baiting or filtering (Table 2). Phytophthora sp. stellaris (4% of the isolates recovered from MP and PMB) and P. sp. Umtamvuna (rarely recovered in UM), both represent novel species residing in ITS Clade 9. Hybrid taxon P. sp. xWS (20% of the isolates) was recovered from PMB and GT and P. sp. xHennops was recovered from UMT, PMB and GT (Table 2, Fig. 1).
Discussion
Fourteen Phytophthora taxa were isolated from soil and water associated with asymptomatic vegetation in natural ecosystems of South Africa. Six of the taxa were of the known species, P. multivora, P. capensis, P. frigida, P. cinnamomi, P. cinnamomi var. parvispora, and P. cryptogea, and three match previously informally designated taxa, P. sp. emzansi, P. sp. PgChlamydo, P.sp. Kununurra. The remaining five taxa did not correspond to any known species. Two of these taxa, found exclusively from water sampling, are thought to be hybrids.
Phytophthora multivora is a species recently described causing disease in natural ecosystems in Western Australia where it has a wide distribution and host range and is a pathogen of Eucalyptus spp., Banksia spp., and Agonis flexuosa (Burgess et al. 2009, Scott et al. 2009). This species was previously misidentified as P. citricola (Burgess et al. 2009) and in Western Australia it has a wider distribution within natural ecosystems than P. cinnamomi (Burgess et al. 2009, Scott et al. 2009). It is also the dominant species in the urban environment on numerous hosts in Myrtaceae and Proteaceae (Barber et al. 2012). The variability within the coxI region led Scott et al. (2009) to hypothesize that P. multivora was endemic to Western Australia. However, similar variability was seen among isolates from South Africa in this study. Additionally, P. multivora was routinely isolated from the rhizosphere of non-symptomatic vegetation in South Africa, while in Australia it is associated with dead or dying vegetation. This is obviously an important species and further studies should be undertaken to determine its origin.
Phytophthora capensis and P. sp. emzansi have been recognized only recently from cultivated endemic plant species in the Western Cape (Bezuidenhout et al. 2010). The recovery of these species in other locations in South Africa shows they have a wider distribution within the region and additional isolates of P sp. emanzsi will facilitate its formal description. Both species are related to P. multivora, and their presence in soil from natural forests where the vegetation was asymptomatic suggests they are probably endemic in South Africa.
Phytophthora frigida was first recovered from diseased roots or the rhizosphere of dying Eucalyptus in South Africa (Maseko et al. 2007). This species was not highly pathogenic to Eucalyptus when compared to P. cinnamomi, a known and serious root pathogen of Eucalyptus in South Africa (Linde et al. 1994). However, it may be a potential threat to other plants in the native vegetation, plantations or in agriculture due to the presence of inoculum in waterways. To date this species has not been recovered elsewhere in the world and it could be endemic to southern Africa. Interestingly, a recently described species from Western Australia, P. elongata, highly pathogenic to young Eucalytus marginata, is the closest relative of P. frigida (Rea et al. 2010).
Since the first report of P. cinnamomi in South Africa in 1933, this species has been the most widely studied Phytophthora species in the country. It is also the most destructive species in native vegetation of the Western Cape Province and in forestry plantations and fruit orchards widely distributed in South Africa (von Broembsen 1984a, 4b, Linde et al. 1994). Thus, the isolation of P. cinnamomi in this study was not surprising. Phytophtora cryptogea, although rarely encountered in this study has also been commonly isolated in agricultural systems in South Africa (Crous et al. 2006, Nagel et al. 2013a).
Phytophthora sp. PgChlamydo has been recovered from stream water, soil, dying plants in many parts of the world including countries of Europe, North and South America, Australia and South Africa (Brasier et al. 2003, Greslebin et al. 2005, Burgess et al. 2009, Reeser et al. 2011, Nagel et al. 2013b, Hüberli et al. 2013). It is morphologically very similar to P. gonapodyides and differs only in its production of chlamydospores. Both species are frequently present in waterways and it is considered a weak pathogen and litter decomposer (Brasier et al. 2003, Jung et al. 2011).
Phytophthora sp. xWS differs from all known species or designated taxa in ITS Clade 6 (Jung et al. 2011). Polymorphism is observed in the ITS sequence data and two separate coxI alleles are obtained, one of which corresponds to P. thermophila and the other corresponds to P. amnicola; both these species have been described recently from Western Australia (Jung et al. 2011, Crous et al. 2012). Phytophthora sp. xWS appears to match a stable hybrid recently characterized from both Australia and South Africa (Hüberli et al. 2013, Nagel et al. 2013b).
One isolate designated as P. sp. canthium was recovered from soil in Umtamvuna Nature Reserve. This isolate is interesting because, based on ITS and cox1 sequence data, it resides in Clade 10. This is a sparsely populated Clade, basal to the Phytophthora phylogeny, presently comprising of only four species, P. morindae, P. kernoviae, P. gallica, and P. boehmeriae. Phytophthora boehmeriae is a species with a global distribution, including South Africa (Roux & Wingfield 1997). The other species in this Clade have a limited geographic distribution, with P. kernoviae being an invasive and damaging pathogen on ornamental and wild plant species in the UK (Brasier et al. 2005).
Four undescribed taxa found in this study reside in ITS Clade 9; one is an exact match for P. sp. Kunnunara, P. sp Umtamvuna is closely related to P. hydropathica and P. sp. stellaris resides in the sub-clade containing P. insolata and P. polonica. The remaining taxon, P. sp. xHennops, appears to be a hybrid; it has two ITS alleles, one of which is an exact match for the type isolate of P. hydropathica. There are also two alleles of the coxI among isolates of P. sp. xHennops (i.e. each isolate only has one coxI allele but was designated as P. sp. xHennops based on having two distinct ITS alleles), one is identical to P. hydropathica, the other to P. sp. Kununurra. Several species belonging to ITS Clade 9 have recently been described from irrigation water (Hong et al. 2008, Hong et al. 2010), however, the pathogenicity of those species is unknown and, like many Clade 6 Phytophthoras, they may be saprophytes of green litter (Brasier et al. 2003, Jung et al. 2011).
Phytophthora sp. stellaris has a single coxI allele, but two ITS alleles. Similarly, P. amnicola returned single alleles for five gene regions, while two ITS alleles, differing by a single indel of 5bp, were obtained (Crous et al. 2012). For P. sp. stellaris, an indel in the ITS allele of some isolates of P. sp. stellaris precluded direct sequencing, however the alleles differed by only by 3–5b p and we consider this to represent intraspecific variation.
This study has contributed considerably to the knowledge of Phytophthora species associated with natural vegetation in South Africa. Given that the relatively few locations considered resulted in a large number of undescribed taxa, additional surveys will undoubtedly reveal an even more substantial Phytophthora biodiversity in South Africa.
Notes
1Informal names have been used in the past for some species discussed here, and also for some that are newly reported. This practice is adopted pending fuller information being obtained, and formal names will be introduced where appropriate in a future publication
References
Barber P, Burgess TI, Paap T, Hardy GESJ (2012) Phytophthora species associated with disease in peri-urban woodland and forest ecosystems. In: 6th International Meeting IUFRO Working Party 7.02.09. Phytophthora in Forests and Natural Ecosystems, 9–14th September Cordoba, Spain: 84.
Bezuidenhout CM, Denman S, Kirk SA, Botha WJ, Mostert L, McLeod A (2010) Phytophthora taxa associated with cultivated Agathosma, with emphasis on the P. citricola complex and P. capensis sp. nov. Persoonia 25: 32–49.
Brasier CM, Beales PA, Kirk SA, Denman S, Rose J (2005) Phytophthora kernoviae sp. nov., an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK. Mycological Research 109: 835–859.
Brasier CM, Cooke DEL, Duncan JM, Hansen EM (2003) Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides-P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycological Research 107: 277–290.
Bumbieris M (1976) The role of Phytophthora cryptogea and waterlogging in a decline of Pinus radiata. Australian Journal of Botany 24: 703–709.
Burgess TI, Wingfield MJ (2001) Exotic pine forestry in the Southern Hemisphere: a brief history of establishment and quarantine practices. South African Forestry Journal 192: 79–84.
Burgess TI, Webster JL, Ciampini JA, White DW, Hardy GESJ, Stukely MJC (2009) Re-evaluation of Phytophthora species isolated during 30 years of vegetation health surveys in Western Australia using molecular techniques. Plant Disease 93: 215–223.
Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology 30: 17–32.
Cooke DEL, Duncan JM (1997) Phylogenetic analysis of Phytophthora species based on ITS1 and ITS2 sequences of the ribosomal RNA gene repeat. Mycological Research 101: 667–677.
Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest Di, Hardy GESJ, Hausbeck MK, Huberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vanky K, Webster BJ, Windstam ST, Groenewald JZ (2012) Fungal Planet description sheets: 107–127. Persoonia 28: 138–182.
Crous PW, Phillips AJL, Baxter AP (2000) Phytopathogenic Fungi from South Africa. Stellenbosch: University of Stellenbosch Printers.
Crous PW, Rong IH, Wood A, Lee S, Glen H, Botha WJ, Slippers B, de Beer WZ, Wingfield MJ, Hawksworth DL (2006) How many species of fungi are there at the tip of Africa? Studies in Mycology 55: 13–33.
Greslebin AG, Hansen EM, Winton LM, Rajchenberg M (2005) Phytophthora species from declining Austrocedrus chilensis forests in Patagonia, Argentina. Mycologia 97: 218–228.
Grundy I, Wynberg R (2001) Integration of biodiversity into national forest planning programmes: the case of South Africa. International Workshop of Integration of Biodiversity into National Forest Planning Programmes. CIFOR Headquarters, Bogor, Indonesia, 13–16 August, 2001: 1–34.
Hong C, Gallegly ME, Richardson PA, Kong P, Moorman GW (2008) Phytophthora irrigata, a new species isolated from irrigation reservoirs and rivers in eastern United States of America. FEMS Microbiology Letters 285: 203–211.
Hong C, Gallegly ME, Richardson PA, Kong P, Moorman GW, LeaCox JD, Ross DS (2010) Phytophthora hydropathica, a new pathogen identified from irrigation water, Rhododendron catawbiense and Kalmia latifolia. Plant Pathology 59: 913–921.
Huberli D, Hardy GES, White D, Williams N, Burgess TI (2013) Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australasian Plant Pathology 42: 251–260.
Hüberli D, Tommerup IC, Hardy GES (2000) False-negative isolations or absence of lesions may cause mis-diagnosis of diseased plants infected with Phytophthora cinnamomi. Australasian Plant Pathology 29: 164–169.
Jung T, Stukely MJC, Hardy GES, White D, Paap T, Dunstan WA, Burgess TI (2011) Multiple new Phytophthora species from ITS clade 6 associated with natural ecosystems in Australia: evolutionary and ecological implications. Persoonia 26: 13–39.
Knox Davies PS (1975) Decline of silver trees and other indigenous species. Veld and Flora 61: 20–22.
Linde C, Kemp GHJ, Wingfield MJ (1994) Pythium and Phytophthora species associated with eucalypts and pines in South Africa. European Journal of Forest Pathology 24: 345–356.
Marincowitz S, Crous PW, Groenewald JZ, Wingfield MJ (2008) Microfungi occurring on the Proteaceae in the Fynbos. [CBS Biodiversity Series no. 7.] Utrecht: CBS-KNAW Fungal Biodiversity Centre.
Martin FN, Tooley PW (2003) Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95: 269–284.
Maseko B, Burgess TI, Coutinho T, Wingfield MJ (2001) First report of Phytophthora nicotianae associated with Eucalyptus die-back in South Africa. Plant Pathology 50: 413.
Maseko B, Coutinho TA, Burgess TI, Wingfield BD, Wingfield MJ (2007) Two new species of Phytophthora from South African eucalypt plantations. Mycological Research 111: 1321–1338.
Nagel JH, Gryzenhout M, Slippers B, Wingfield MJ (2013a) The occurrence and impact of Phytophthora on the African continent. In: Phytophthora Diseases: a global perspective (Lamour K, ed.): pp. 204–214. Wallingford: CAB International.
Nagel JH, Gryzenhout M, Slippers B, Wingfield MJ, Hardy GESJ, Stukely MJC, Burgess TI (2013b) Characterization of Phytophthora hybrids 1 from ITS clade 6 associated with riparian ecosystems in South Africa and Australia. Fungal Biology. 10.1016/j.funbio.2013.1003.1004
Oudemans PV, Förster H, Coffey MD (1994) Evidence for distinct isozyme subgroups within Phytophthora citricola and close relationships with P. capsici and P. citrophthora. Mycological Research 98: 189–199.
Posada D (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
Rea A, Jung T, Burgess TI, Stukely MJC, Hardy GESJ (2010) Phytophthora elongata sp. nov. a novel pathogen from the Eucalyptus marginata forest of Western Australia. Australasian Plant Pathology 39: 477–491.
Reeser PW, Sutton W, Hansen EM, Remigi P, Adams GC (2011) Phytophthora species in forest streams in Oregon and Alaska. Mycologia 103: 22–35.
Robideau GP, de Cock AWAM, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Desaulniers N, Eggertson QA, Gachon CMM, Hu C-H, Küpper FC, Rintoul TL, Sarhan E, Verstappen ECP, Zhang Y, Bonants PJM, Ristaino JB, Lévesque CA (2011) DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 11: 1002–1011.
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Roux J, Wingfield MJ (1997) Survey and virulence of fungi occurring on diseased Acacia mearnsii in South Africa. Forest Ecology and Management 99: 327–336.
Scott PM, Burgess TI, Barber PA, Shearer BL, Stukely MJC, Hardy GESJ, Jung T (2009) Phytophthora multivora sp. nov, a new species recovered from declining Eucalyptus, Banksia, Agonis and other plant species in Western Australia. Persoonia 22: 1–13.
von Broembsen S (1984a) Distribution of Phytophthora cinnamomi in rivers of the southwestern Cape province. Phytophylactica 16: 227–229.
von Broembsen S (1984b) Occurrence of Phytophthora cinnamomi on indigenous and exotic hosts in South Africa, with special reference to the southwestern Cape Province. Phytophylactica 16: 221–225.
Von Broembsen SL, Kruger FJ (1985) Phytophthora cinnamomi associated with mortality of native vegetation in South Africa. Plant Disease 69: 715–717.
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (MA Innes, DH Gelfand, JJ Sninsky, TJ White (eds): 315–322. San Diego: Academic Press.
Wingfield MJ, KnoxDavies PS (1980) Observations on diseases in pine and eucalyptus plantations in South Africa. Phytophylactica 12: 57–63.
Winton LM, Hansen EM (2001) Molecular diagnosis of Phytophthora lateralis in trees, water, and foliage baits using multiplex polymerase chain reaction. Forest Pathology 31: 275–283.
Zeijlemaker FCJ (1971) Black-butt disease of black wattle caused by Phytophthora nicotianae var. parasitica. Phytopathology 61: 144–145.
Acknowledgements
We are grateful to the Department of Science and Technology (DST)/National Science Foundation (NRF) Centre of Excellence in Tree Health Biotechnology (CTHB) for financial assistance to undertake to study. We also acknowledge support from the University of Pretoria that provided a post-doctoral fellowship for E. O. Various students and colleagues provided technical support, particular regarding the collections of samples and we specially acknowledge Jan Nagel and Hugh Glen in this regard. We kindly thank Chuan Hong and Patricia Richardson for providing coxI sequence data for several Phytophthora aquimorbida, P. hydropathica, P. irrigate, and P. parsiana.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Oh, E., Gryzenhout, M., Wingfield, B.D. et al. Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus 4, 123–131 (2013). https://doi.org/10.5598/imafungus.2013.04.01.12
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.5598/imafungus.2013.04.01.12