Abstract
The Pinus mugo complex comprises closely related pines in different taxonomic ranks, which commonly occur in major European mountain chains. This large and polymorphic complex has been, and still is, a considerable challenge for taxonomists due to highly variable phenotypical characteristics within particular taxa, ongoing hybridization in sympatric populations, lack of distinct morphological species-specific determinants, and the absence of distinct genetic differentiation among them. Moreover, cytogenetic data on pines from this complex are very scarce. Therefore, in this study, we used C-banding methods and flow cytometric analysis in order to provide new data on karyotypical features and DNA content of three closely-related pines from the Pinus mugo complex, i.e., Pinus mugo (Turra), Pinus uliginosa (Neumann), and Pinus × rhaetica (Brügger) and to gain more insight into the taxonomic relationships between these pines. Our results confirmed that the basic number of chromosome is the same amongst the analyzed taxa (2n = 2x = 24), and that the karyotypes have a similar morphology. The number of secondary constrictions (SCs) and C-banding patterns differed slightly between P. mugo, P. uliginosa and P. × rhaetica. The nuclear DNA content (2C/pg) among analyzed taxa is from 46.41 pg for P. mugo and P. × rhaetica to 46.48 pg for P. uliginosa. For each of these pines, certain taxa-specific cytogenetic determinants were identified. Our results indicate that they may represent two different taxa, and so their names should not be used interchangeably in scientific literature.
Similar content being viewed by others
Introduction
The aggregate of closely-related European mountain pines, known as the Pinus mugo complex, comprises about one hundred forms (Christensen 1987). Pines belonging to this complex play an important ecological role and grow in the major European mountain chains, including the Pyrenees, the Alps, and the Carpathians (Hamerník and Musil 2007). The most well-known and most widely studied taxa from the Pinus mugo complex are Pinus mugo Turra (dwarf mountain pine), Pinus uliginosa Neumann (peat-bog pine) and Pinus uncinata Ramond (mountain pine).
The relationships among these taxa and potential species-specific determinants for them have been investigated extensively using comparative analysis on needle traits and cones (Boratyńska and Boratyński 2007), the serological methods (Prus-Głowacki et al. 1985), allozymes (Prus-Głowacki et al. 1998; Lewandowski et al. 2000), the chemotaxonomic approach (Bonikowski et al. 2015; Celiński et al. 2015), or – more recently – using DNA markers (Monteleone et al. 2006; Celiński et al. 2017a) and next-generation sequencing (NGS) (Celiński et al. 2017b).
Despite many investigations and the variety of applied techniques, the origin, species distinctiveness, and relationships among the taxa as well as their taxonomic status within the Pinus mugo complex are still enigmatic. Moreover, some of the taxa included in this complex, such as P. uliginosa, have many synonymous names in literature. Some researchers identify P. uliginosa as P. rotundata, while other identify it as Pinus × rhaetica (Christensen 1987; Hamerník and Musil 2007). In Poland the peat-bog pine is a highly endangered and protected taxon occurring only in a few small populations. Correct nomenclature and identification are crucial for successful conservation of this pine.
Cytogenetic techniques are very useful for the study of phylogenetic and systematic relationships between pine species (Bogunić et al. 2011). There are many karyological reports for different pine species, which were obtained using conventional (Mehes-Smith et al. 2011) and molecular cytogenetic methods (Hizume et al. 2002; Nkongolo and Mehes-Smith 2012). Some data exist concerning the taxa from the Pinus mugo complex (Bogunić et al. 2011), but no-one has investigated P. uliginosa or P. × rhaetica yet.
Phylogenetic results as well as systematic relationships can be complemented with information obtained using flow cytometry analysis of DNA content. Genome size data can be used not only for taxonomic purposes as a species-specific marker, but also to understand the dynamics of genome evolution (Loureirol et al. 2010).
The aim of this study was to apply C-banding methods and flow cytometric analysis in order to provide new data on karyotypical features and DNA content of three closely-related pines from the Pinus mugo complex and to gain more insight into the taxonomic relationships between these pines. This is also the first report on DNA content and karyotypes for Pinus uliginosa and Pinus × rhaetica.
Material and methods
Plant material
This study used the needles and seedlings of four pines, i.e., P. mugo, P. uliginosa, and P. × rhaetica from the Pinus mugo complex and Pinus sylvestris as the outer group. Seedlings were used for cytological research, whereas fresh needles were used for DNA content analyses. The needles and seeds of P. mugo, P. uliginosa and P. × rhaetica were collected in the Dendrological Garden University of Life Sciences in Poznań. Individuals of Pinus mugo, Pinus uliginosa and P. × rhaetica growing there, represent natural Polish populations of these taxa located in the Tatras, the Węgliniec Nature Reserve and the peat bog “Bór na Czerwonym” Nature Reserve, respectively. The needles and seeds of P. sylvestris were collected from a natural stand near Collegium Biologicum of Adam Mickiewicz University, Poznań.
Flow cytometry
Nuclear genome was determined using a Cytomics FC500 (Beckman Coulter) flow cytometer with a 488 nm laser. Pine needles were washed three times in distilled water. The needles were simultaneously chopped with a 1 cm2 leaf of Vicia faba L. “Inovec” (internal standard, 2C = 26.90 pg) using ice-cold Galbraith’s buffer (Galbraith et al. 1983) containing 0.1% Triton, 1% (w/v) polyvinylpyrrolidone 40,000 and 5 μl/ml of 2-mercaptoethanol. The suspensions were filtered through a 30 μm nylon mesh (CellTrics, Sysmex Partec), and 50 μl/ml RNAse was added. Finally, nuclei were stained with 50 μl/ml propidium iodide and incubated on ice for 10–15 min. Five specimen of each species were measured separately with three repetitions – at least 5000 nuclei were analyzed. The DNA amount in each pine species was calculated based on values of G1 peak means of samples and the internal standard.
Aceto-orceine staining, C-banding and karyotype analysis
Root tips were obtained from seedlings germinated on moist filter paper in glass Petri dishes. When the seedling roots reached a length of 2 to 3 cm, they were pre-treated with 0.05% colchicine for 16 h at room temperature, then fixed in a 3: 1 mixture of 95% ethanol and glacial acetic acid at 4 °C for a minimum of 48 h. For the purposes of karyotype observation, chromosomes were stained in aceto-orceine after hydrolysis in 1 N HCl at 60 °C for 12 min. The C-banding protocol described by Mehes-Smith et al. (2011) was followed. The arm lengths of chromosomes in the CCD-captured images of ten cells from each of two or three chromosome spreads of each species were measured. The arm ratio was calculated, and the average values were used to construct an ideogram of the karyotype for each species. The chromosomes were assigned consecutively according to their lengths, with the longest being assigned as I and the shortest as XII. For chromosomes of equal length, the chromosome with the longest short arm was assigned first. In addition to the length and arm ratios of the chromosomes, secondary constrictions (SCs) and C-banding patterns were also shown on the ideograms.
Results and discussion
Genome size is important in the context of possible taxonomical relationship among the closely-related taxa of pine species, especially in combination with quantitative (chromosome length, asymmetric indices) and qualitative (C-banding) cytogenetic data. 2C DNA content is considered to be a useful parameter in different areas of species biology (Bennett and Leitch 2011; Sedelnikova 2016). According to Sedelnikova (2016) the variability in the content of DNA in the genus Pinus is related to its taxonomic position.
Using nuclei isolated from fresh needles together with a leaf of Vicia faba as an internal standard, we estimated a 2C DNA content as an average from 10 flow-cytometric measurements in the amounts of 46.3 ± 0.12 pg for P. sylvestris and 46.41 ± 0.23 pg for P. mugo (Fig. 1). These values are higher than in previous reports for P. mugo: 42.56 ± 0.79, ranging from 41.08 to 43.95 pg (Morse et al. 2009) and lower for P. sylvestris: 49.25 ± 2.65 pg (Valkonen et al. 1994). However, it is assumed that growth conditions, especially altitude, can change the DNA content in the nuclear genome of plants (Sedelnikova 2016). The impact of this factor can be ruled out in this study due to the lack of significant differences in altitude between the analyzed samples. The DNA contents of P. uliginosa (46.48 ± 0.18 pg) and P. × rhaetica (46.41 ± 0.22 pg) have been determined for the first time. Experimental data suggest that hybridization in pines caused stability in nuclear DNA content (Williams et al. 2002). The hybrid genomes of P. uliginosa and P. × rhaetica possess neither increased nor decreased DNA content, irrespective of the phylogenetic distance between the suggested parents, P. sylvestris and P. mugo (Christensen 1987; Lewandowski and Dering 2006).
The lack of a difference in genome size does not exclude substantial differences in the structure of the karyotype. In the case of European black pine (Pinus nigra Arn.), despite no differences in 2C DNA, a significant variation among karyotypes studied using fluorescence in situ hybridization of 5S rDNA and 18S rDNA was observed (Nkongolo and Mehes-Smith 2012).
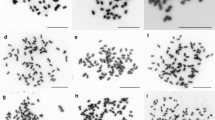
Our results confirmed that basic chromosome number is the same among the studied taxa (2n = 2x = 24), and that the karyotypes have a similar morphology. We analyzed numerical parameters, such as chromosome length, centromeric index, or asymmetrical indices (Table 1). We used these data to compare interspecific karyotypes. Chromosomes type were defined based on the centricity class values, taking into account only: M, metacentric (CT ± 47); NSM, near submetacentric (42 ± CT < 47) and SM, submetacentric (CT < 42) (Guerra 1986). Secondary constrictions (SCs) were observed in the majority of the analyzed material, and their number seemed to be one of the most important karyological features which differ the studied taxa. The highest number of SCs was observed in P. × rhaetica (8). For P. mugo and P. uliginosa, an equal number of SCs was noted (5), while in P. sylvestris, we observed 7 SCs. A Giemsa C-banding analysis revealed a slight difference in the distribution of C-heterochromatin between the studied taxa (Fig. 2). We confirmed that a cytologically-detectable heterochromatin was found in the vicinity of centromeres in all karyotypes. Such pericentromeric C-heterochromatin is characteristic for the subgenus Pinus (Hizume et al. 2002). All four species contained distally located C-heterochromatin with 9 to 16 intercalary C-bands per haploid genome. These intercalary C-bands were observed as a single narrow band restricted to one arm of a banded chromosome. The taxa P. × rhaetica, P. uliginosa and P. mugo showed 3 to 8 chromosomes with two C-bands, one on each arm (Fig. 2 b, c, d).
Many studies report a lack of correlation between cytological and molecular evolution. For example, the classification of the section Pinus based on FISH karyotyping corresponds with the morphology-based classification of Price et al. (1998), but is incompatible with the taxonomy of Gernandt et al. (2005), which was based on chloroplast DNA. There are no such data available for the Pinus mugo complex.
Our studies confirm differences in the distribution of cytologically detectable heterochromatin and the secondary constructions (SCs) among the studied taxa, but the most visible differences apply to P. mugo – P. sylvestris chromosomes. The observed variation of karyotypes is inconsistent with observations at the DNA level. According to Celiński et al. (2017b) it is not possible to distinguish particular pine taxa inside the Pinus mugo complex using chloroplast barcodes due to lack of differences in DNA sequences. The deficiency of genetic differences in the obtained chloroplast DNA sequences may result from recent diversification or hybridisation inside these taxa. Additional analyses that would combine different molecular and cytogenetic techniques may validate the association between cytological and molecular data in the Pinus mugo complex.
Conclusions
The obtained results indicate that Pinus uliginosa and Pinus × rhaetica show certain taxa-specific determinant connected with C-banding patterns, but insufficient levels of variation in the banding patterns made it difficult to unambiguously discriminate among most chromosomes.
In our opinion these pines represent two different taxa, and so their names should not be used as synonyms, but additional analyses that would combine different molecular and cytogenetic techniques are necessary to be able to clearly determine the taxonomic status of pines belonging to the Pinus mugo complex.
References
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107:467–590. https://doi.org/10.1093/aob/mcq258
Bogunić F, Siljak-Yakovlev S, Muratović E, Pustahija F, Medjedović S (2011) Molecular cytogenetics and flow cytometry reveal conserved genome organization in Pinus mugo and P. uncinata. Ann For Sci 68:179–187. https://doi.org/10.1007/s13595-011-0019-9
Bonikowski R, Celiński K, Wojnicka-Półtorak A, Maliński T (2015) Composition of essential oils isolated from the needles of P. uncinata Ramond ex. Mirb. And P. uliginosa Neumann ex Wimmer grown in Poland. Nat Prod Commun 10:371–373
Boratyńska K, Boratyński A (2007) Taxonomic differences among closely related pines Pinus sylvestris, P. mugo, P. uncinata, P. rotundata and P. uliginosa as revelated in needle sclerenchyma cells. Flora 202:555–569. https://doi.org/10.1016/j.flora.2006.11.004
Celiński K, Bonikowski R, Wojnicka-Półtorak A, Chudzińska E, Maliński T (2015) Volatiles as chemosystematic markers for distinguishing closely related species within the Pinus mugo complex. Chem Biodivers 12:1208–1213. https://doi.org/10.1002/cbdv.201400253
Celiński K, Kijak H, Barylski J, Grabsztunowicz M, Wojnicka-Półtorak A, Chudzińska E (2017a) Characterization of the complete chloroplast genome of Pinus uliginosa (Neumann) from the Pinus mugo complex. Conserv Genet Resour 9:209–212. https://doi.org/10.1007/s12686-016-0652-6
Celiński K, Kijak H, Wojnicka-Półtorak A, Buczkowska-Chmielewska K, Sokołowska J, Chudzińska E (2017b) Effectiveness of the DNA barcoding approach for closely related conifers discrimination: A case study of the Pinus mugo complex. C R Biol 340:339–348. https://doi.org/10.1016/j.crvi.2017.06.002
Christensen KI (1987) Taxonomic revision of the Pinus mugo complex and P. × rhaetica (P. mugo × P. sylvestris) (Pinaceae). Nord J Bot 7:383–408. https://doi.org/10.1111/j.1756-1051.1987.tb00958.x
Galbraith D, Harkins K, Maddox J, Ayres N, Sharma D, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:1049–1051. https://doi.org/10.1126/science.220.4601.1049
Gernandt DS, López GG, García SO, Liston A (2005) Phylogeny and classification of Pinus. Taxon 54:29–42. https://doi.org/10.2307/25065300
Guerra MDS (1986) Reviewing the chromosome nomenclature of Levan et al. Braz J Genet 9:741–743
Hamerník J, Musil I (2007) The Pinus mugo complex − its structuring and general overview of the used nomenclature. J For Sci 53:253–266. https://doi.org/10.17221/2020-JFS
Hizume M, Shibata F, Matsusaki Y, Garajova Z (2002) Chromosome identification and comparative karyotypic analyses of four Pinus species. Theor Appl Genet 105:491–497. https://doi.org/10.1007/s00122-002-0975-4
Lewandowski A, Dering M (2006) Crossability between Pinus uliginosa and its putative parental species Pinus sylvestris and Pinus mugo. Silvae Genet 55:52–54. https://doi.org/10.1515/sg-2006-0008
Lewandowski A, Boratyński L, Mejnartowicz LE (2000) Allozyme investigations on the genetic differentiation between closely related pines − Pinus sylvestris, P. mugo, P. uncinata and P. uliginosa (Pinaceae). Plant Syst Evol 21:15–24. https://doi.org/10.1007/BF01086377
Loureirol J, Trávníček P, Rauchová J, Urfus J, Vít P, Štech M, Castrol S, Suda J (2010) The use of flow cytometry in the biosystematics, ecology and population biology of homoploid plants. Preslia 82:3–21
Mehes-Smith M, Nkongolo KK, Kim NS (2011) A comparative cytogenetic analysis of five pine species from North America, Pinus banksiana, P. contorta, P. monticola, P. resinosa, and P. strobus. Plant Syst Evol 292:153–164. https://doi.org/10.1007/s00606-010-0401-3
Monteleone I, Ferrazzini D, Belletti P (2006) Effectiveness of neutral RAPD markers to detect genetic divergence between the subspecies uncinata and mugo of Pinus mugo Turra. Silva Fenn 40:391–406. https://doi.org/10.14214/sf.476
Morse AM, Peterson DG, Islam-Faridi MN, Smith KE, Magbanua Z, Garcia SA, Kubisiak TL, Amerson HV, Carlson JE, Nelson D, Davis JM (2009) Evolution of genome size and complexity in Pinus. PLoS One 4(4332). https://doi.org/10.1371/journal.pone.0004332
Nkongolo KK, Mehes-Smith M (2012) Karyotype evolution in the Pinaceae: implication with molecular phylogeny. Genome 55:735–753. https://doi.org/10.1139/g2012-061
Price RA, Liston A, Strauss SH (1998) Phylogeny and systematics of Pinus. In: Richardson DM (ed) Ecology and biogeography of Pinus. Cambridge University Press, London, pp 49–68
Prus-Głowacki W, Szweykowski J, Nowak R (1985) Serotaxonomical investigation of the European pine species. Silvae Genet 34:162–170
Prus-Głowacki W, Bujas E, Ratyńska H (1998) Taxonomic position of Pinus uliginosa Neumann as related to other taxa of Pinus mugo complex. Acta Soc Bot Pol 67:269–274. https://doi.org/10.5586/asbp.1998.035
Sedelnikova TS (2016) Variability of genome size in conifers under extreme environmental conditions. Biol Bull 6:177–188. https://doi.org/10.1134/S2079086416020079
Valkonen JPT, Nygren M, Ylonen A, Mannonen L (1994) Nuclear DNA content of Pinus sylvestris (L.) as determined by laser flow cytometry. Genetica 92:203–207. https://doi.org/10.1007/BF00132539
Williams CG, Joyner KL, Auckland LD, Johnston S, Price HJ (2002) Genomic consequences of interspecific Pinus spp. hybridization. Biol J Linn Soc 75:503–508. https://doi.org/10.1046/j.1095-8312.2002.00038
Acknowledgments
This research was financially supported by the Ministry of Science and Higher Education in Poland (No. NN304060339).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Celiński, K., Chudzińska, E., Gmur, A. et al. Cytological characterization of three closely related pines - Pinus mugo, P. uliginosa and P. × rhaetica from the Pinus mugo complex (Pinaceae). Biologia 74, 751–756 (2019). https://doi.org/10.2478/s11756-019-00201-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-019-00201-6