Abstract
Purpose
Sarcocystis spp. are protozoan parasites of livestock which also infect birds, lower vertebrates and mammals, including man. Wild and domestic ruminants such as red deer, roe deer, fallow deer, cattle, sheep and goats may act as intermediate hosts for many Sarcocystis species, some of which are significant pathogens causing sarcocystosis in livestock and humans. The purpose of the present study was to determine the prevalence of Sarcocystis species in fallow deer farmed in an open pasture system.
Methods
Samples of heart and oesophagus tissue taken from five fallow deer were examined by light microscope for the presence of sarcocysts. Genomic DNA was extracted from individual sarcocysts. ssu rRNA was successfully amplified using their DNA as templates.
Results
Analysis of the ssu rRNA identified the presence of two S. morae sarcocysts in the heart tissue; similarly, S. gracilis sarcocysts were identified in the heart and oesophagus, and Sarcocystis sp. most closely related to S. linearis and S. taeniata were detected in oseophagus.
Conclusions
These findings confirm the presence of Sarcocystis spp. in farmed fallow deer in Poland; however, more molecular studies are needed.
Similar content being viewed by others
Introduction
Sarcocystis spp. are obligate heteroxenous protozoan parasites (Apicomplexa, Sarcocystidae): the continuation of their life cycle requires the presence of both intermediate and definitive hosts. The intermediate host, which is usually a herbivore or omnivore infected by ingesting food or water contaminated with sporocysts, hosts the asexual phase of the life cycle. In immature cysts, further asexual reproduction takes place by repeated endodyogeny of metrocytes. Mature cysts contain several hundred thousand bradyzoites; these do not divide further and represent the terminal asexual stage in the intermediate host. Following the ingestion of their cysts by a carnivorous definitive host, the bradyzoites initiate the sexual phase of the life cycle (gamogony) in the cells of the small intestine of the host, ultimately resulting in the formation of oocysts [1]. Free sporocysts are released into the intestinal lumen and passed into the soil or water of the surrounding environment with the faeces.
Sarcocystosis is a zoonotic disease presented by a wide range of domestic ungulates, including cattle, sheep, goats or water buffalo, or wild ones, such as camels and wild boar [2,3,4,5,6]. The Sarcocystis of cervids has been intensively studied over the last few years. At least 14 Sarcocystis species have so far been identified in red deer (Cervus elaphus) in Europe: S. cervicanis, S. elongata, S. cf. grueneri, S. hardangeri, S. hjorti, S. cf. hofmanni, S. iberica, S. linearis, S. morae, S. ovalis, S. taeniata, S. tarandi, S. truncata and S. venatoria [3, 4, 7, 8]; however, little is known of their potential zoonotic or public health significance [9, 10]. Although more than 200 valid Sarcocystis species have been identified, only three, S. hominis, S. suihominis, and S. heydorni, have so far been shown to be capable of causing intestinal disease in humans [4, 11]. Although the intestinal forms of infection are often asymptomatic and self-limiting, a number of studies have reported symptoms ranging from mild gastrointestinal distress to nausea, loss of appetite, vomiting, stomach ache, bloat, diarrhea, dyspnea and tachycardia [12]. Whether individuals remain asymptomatic or develop some degree of disease appears to be related to the quantity of the meat consumed, inoculum size and various host factors [12]. Compared to red deer, less data concern on fallow deer (Dama dama) [13,14,15,16,17,18]. The latest work [19] report fallow deer in Spain as a new host for S. morae, originally described from the red deer. Despite being regularly consumed in other parts of the EU, meat from fallow deer and red deer is just entering the market for human consumption in Poland. It is important to note that although the numbers of wild and farmed fallow deer have increased over the last decade in the country, no studies have yet examined the presence of Sarcocystis species among them. The present paper, therefore, aims to determine the presence of Sarcocystis species in fallow deer farmed in an open pasture system.
Materials and methods
Animals and sampling
The study was carried out in the Breeding Station of the Witold Stefański Institute of Parasitology, Academy of Sciences in Kosewo Górne (Region of Warmia and Mazury; Poland; N: 53° 48′; E: 21° 71 23′). Samples of heart and oesophagus tissue were obtained from five 1-year-old male fallow deer slaughtered for commercial purposes. All samples (10 g of weight) were gently broken up in physiological saline solution using a blender and filtered through gauze and the resulting sediments were collected. The basic morphology (shape, size) of any cysts present was examined in wet mounts under an inverted Olympus IX50 microscope fitted with a camera. The samples were preserved for molecular investigation in sterile H2O in Eppendorf tubes and stored at − 72 °C.
DNA extraction
Individual sarcocysts were detected in muscles by light microscopy. Total DNA was extracted from 10 individual sarcocysts using the NucleoSpin®Tissue kit (Macherey–Nagel, Germany) according to the manufacturers’ instructions. The DNA was eluted in 50 μl of distilled water. The extracted DNA was stored at − 20 °C for PCR assay.
Two different PCR protocols were employed for the molecular examination of tissue cysts
In the first protocol, DNA amplification was performed using PCR targeted at a 18S rDNA sequence of approximately 900 bp in accordance with More et al. [20]. The following specific primers were used: SarcoFext (GGT GAT TCA TAG TAA CCG AAC G) and SarcoRext (GAT TTC TCA TAA GGT GCA GGA G).
Reactions were performed in a final volume of 25 µl reaction mixture with 1 unit of Taq DNA polymerase per reaction (Thermo Fisher Scientific) under the following conditions (final concentrations): 1× reaction buffer supplied with the DNA polymerase, 1.25 mM MgCl2, 200 mM dNTPs, 0.2 M of each primer and the following cycler program: initial denaturation (95 °C, 5 min), followed by 40 cycles of denaturation (94 °C, 40 s), annealing (59 °C, 1 min), elongation (72 °C, 1 min) and a final extension at 72 °C (10 min).
The PCR products were analyzed by electrophoresis in 1.5% agarose gel and stained with GelRed (Nucleic Acid Gel Stain, Biotium). PCR products 900 bp in size were cut out from the gel and purified using Clean-up Product Purification Kits (A&A Biotechnology, Poland) according to the manufacturer’s instructions. DNA concentration was estimated using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, USA).
The PCR amplicons were then ligated into pGEM-T easy cloning vector (Promega). Escherichia coli strain XL-1 Blue MRF electrocompetent cells (Promega) were used for cloning. Positive clones (six) were identified by colony PCR with primers directed against vector sequences outside the multi-cloning site. The clones found to contain inserts were used for further examination. Positive plasmids were purified using GeneAll Exprep Plasmid SV mini (GeneAll, Korea) according to the manufacturer’s instructions.
The ssu rRNa gene products concentration was measured using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, USA) and was then sequenced (Genomed, Poland). The sequence information obtained from all the isolated clones was assembled using Vector NTI Advance 10 software (Invitrogen, Scotland). The complete sequences were checked against sequences published in GenBank using BLAST (https://www.ncbi.nlm.nih.gov/BLAST/).
In the second protocol, the ssu rRNA gene was amplified by PCR using the primers for Sarcocystis [ERIB1 5′-ACC TGG TTG ATC CTG CCA G-3′, Primer1L 5′-CCA TGC ATG TCT AAG TAT AAG C-3′, Primer3H 5′-GGC AAA TGC TTT CGC AGT AG-3′] [21] in a 25 μl final reaction volume with one unit of Taq DNA polymerase/reaction (Thermo Fisher Scientific). The following final concentrations of reagents were used: 1× reaction buffer, 1.5 mM MgCl2, 250 μM dNTPs, 0.5 μM of each primer. PCR was started with initial denaturation (95 °C, 5 min), followed by 35 cycles of denaturation (95 °C, 30 s), annealing (55.5 °C, 30 s), elongation (72 °C, 1 min) and a final extension at 72 °C (10 min).
PCR products were analyzed and sequenced as described above. The purified products were then sequenced (Genomed, Poland).
Phylogenetic analyses
The evolutionary history was inferred using the Maximum Likelihood method based on the Kimura 2-parameter model [22]. The tree with the highest log likelihood (− 2638.89) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. A discrete Gamma distribution was used to model evolutionary rate differences among sites [four categories (+G, parameter = 0.3385)]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 100 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 789 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [23].
Results
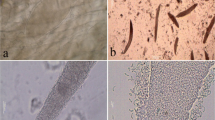
Microscopic sarcocysts were detected in all of the examined fallow deer under light microscope examination in wet mounts. This preliminary microscopic observation revealed the presence of at least three morphs among the isolated sarcocysts depending on their size or shape (Fig. 1a, b).
Two fragments of 18S rDNA (851 bp and 776 bp) were amplified using primers specific for Sarococystis spp. The two nucleotide sequences obtained from sarcocysts isolated from the heart were deposited in GenBank under accession numbers KX364266.1 and KX364267.1. A BLAST-N search found that one nucleotide sequence deposited under accession number KX364266.1 share 99.41% similarity with sequences deposited in GenBank under accession numbers MK790239, MK790238, KY973379.1, KY973375.1 and KY973374.1 and described as S. morae. Second nucleotide sequence deposited under accession number KX364267.1 share 99.74% identity with sequences KY973379.1, KY973375, MK790239 and MK790238—originally described from red deer and newly described in fallow deer.
The primers specific for Sarococystis spp. used in the second PCR protocol were then used to amplify ssu rRNA fragments about 1 kb in length. Three nucleotide sequences of the ssu rRNA were obtained from three sarcocysts: one isolated from fallow deer heart tissue and two isolated from oesophagus tissue. All three sequences were deposited in GenBank under accession numbers MH221019.1 (1064 bp), MH221020.1 (1016 bp) and MH221021.1 (807 bp).
BLAST-N search found nucleotide sequence MH221019.1 obtained from the heart tissue to share 99.91% identity with those from the diaphragm of Capreolus capreolus and deposited in GenBank as S. gracilis under accession numbers KY019031.1 and KY019030.1. Of the nucleotide sequences isolated from the oesophagus, the first (MH221020.1) demonstrated 100% similarity with sequences KY019031.1 and JN256131.1, previously isolated from the diaphragm of Capreolus capreolus and described as S. gracilis. The second sequence (MH221021.1.) represent Sarcocystis sp. and had 96.58%-99.50% identity with S. linearis and 97.09–99.01% identity with S. taeniata.
The phylogenetic analysis placed two new sequences, MH221019.1 and MH221020.1, within the same clade as S. gracilis sequences KY019031.1, KY019030.1, KF880741.1, JN256131.1 and JN226126.1 (Fig. 2). Sequence Sarcocystis sp. (MH221021.1) is closely related to S. linearis (KY973359.1) and S. taeniata (KF831286.1). Further investigations using cox1 are needed for the conclusive identification of MH221021.1. Sequences KX364266.1 and KX364267.1 were placed in the same clade as Sarcocystis sp. sequence KT873778.1 and S. morae sequences (MK790239.1 and MK790238.1) both isolated form sarcocysts from the tongue of fallow deer as well as KY973373.1 and KY973378.1 sequences: these were identified from bradyzoites isolated from the oesophagus of Cervus elaphus.
Phylogenetic tree of Sarcocystidae based on ssu rRNA gene sequences and inferred using the maximum likelihood (ML) method. The ssu rRNA tree was constructed based on the alignment of nearly complete ssu rRNA gene sequences of five Polish Sarcocystis spp. isolates and available ssu rRNA gene sequences of related species deposited in GenBank. The trees were rooted with Eimeria adeneodei and Neospora caninum ssu rRNA sequences. Polish isolates are marked in green (color figure online)
Discussion
Over the last decades, an increasing amount of research has been devoted to the life cycle, morphology and pathogenicity of Sarcocystis species. Although Sarcocystis infection is common in many domestic and wild animal species, little is known about its occurrence in Poland, with only a handful of studies having been performed. One study based on trichinoscopy and the compression method identified the presence of large-scale invasions of cysts in muscle tissue among 166 wild boars, 53 deer and 53 roe deer in the years 1997–99 [2]. A later study of four Polish roe deer based on light microscopy and sequence analysis of small subunit ribosomal RNA (ssu rRNA) and subunit I of cytochrome oxidase (cox 1) identified the presence of S. gracilis, S. oviformis and S. silva sarcocysts [24]. S. tenella has been detected in Tatra chamois (Rupicapra rupicapra tatrica) previously only reported in sheep [25]. A light microscopy study by Pyziel and Demiaszkiewicz (2009) [26] identified S. cruzi in heart, oesophagus and diaphragm muscle from one European bison from the Białowieża Forest.
Although Sarcocystis spp. have been identified in fallow deer in previous studies [13, 14, 17], the precise species was unknown. Only Hernandez-Rodriquez et al. (1992) [15] described S. jorrini sp. nov. as a new species in fallow deer: macroscopic cysts were detected in the skin, and virtually all striated muscles, including those of the esophagus and heart. The cysts themselves were white and spindle shaped, and could clearly be distinguished from the surrounding muscle tissue. Wesemeier and Sedlaczek (1995a) [16] describe the transmission electron microscope identification of sarcocysts in two free-ranging indigenous fallow deer, Dama d. dama (L.), from northeast Germany, and one in captive Persian fallow deer (Dama dama mesopotamica) from the Berlin-Friedrichsfelde Zoo. In the free-ranging D. d. dama, two Sarcocystis species were found to be present: one known and the other new to the host. The known species was also found in D. dama mesopotamica. Based on a subsequent review of previous studies and their own research on roe deer, red deer and wapiti, Wesemeier and Sedlaczek [27], propose that the two Sarcocystis species should be designated S. cf. grueneri (known in D. dama but not named) and S. cf. hofmanni (new for D. dama).
Our results confirm the presence of Sarcocystis spp. in all tested young fallow deer and suggest that a single year of stay in the pastures was sufficient for the animals to become infected. It is important to note that identified Sarcocystis species (S. morae, S. gracilis and Sarcocystis sp.) have not previously been reported in fallow deer in Poland. Lately, S. morae was reported in fallow deer as a new host in Spain by De las Cuevas et al. [19]. Sarcocystis gracilis is considered to be parasites of roe deer (Capreolus capreolus) [28] while S. morae occur in red deer (Cervus elaphus) [8] and fallow deer (Dama dama) [19]. The Sarcocystis isolates obtained in the present study shared 96.58–100% identity with those taken previously from different tissues of various hosts from other geographical localities.
Although Sarcocystis spp. can be recognized using simple and inexpensive methods, such as naked eye examination or light microscopy, a variety of molecular methods have been developed and implemented to identify Sarcocystis spp. and to assess the genetic diversity among these parasite species. The slowly-evolving small subunit (ssu) rRNA gene, commonly used in phylogenetic studies, is also well suited for studying the phylogeny of Sarcocystis species [21, 24, 29, 30]. The phylogenetic tree created from the S. gracilis, S. linearis and Sarcocystis sp. sequences from the fallow deer in the present analysis is in agreement with those described in other studies. Two new S. gracilis sequences (MH221019.1 and MH221020.1), two new S. morae sequences (KX364266.1 and KX364267.1) and one Sarcocystis sp. sequence (MH221021.1) were identified and positioned within clades corresponding to other sequences of the same species. BLAST searches and phylogenetic analyses based on ssu rRNA gene sequences confirmed that our isolates closely matched the sequences of those deposited in GenBank and demonstrated a low level of intraspecific sequence variation. Although Januškevičius et al. [31] conclude that no statistically significant differences in infection prevalence exist between muscle groups in the Cervidae, e.g. sika deer, elk, red deer and roe deer, Malakauskas and Grikienienė [32] found infection intensity to be higher in samples of oesophagus and heart muscle than those from the diaphragm. In the present study, samples were only obtained from oesophagus and heart tissue. Therefore, further studies are needed to determine the presence and the intensity of Sarcocystis spp. infection in other groups of muscles.
Although approximately 200 species of Sarcocystis are recognized, the definitive and intermediate hosts are known for only 62 of them [4]. In the case of S. gracilis, the definitive hosts are known to be dogs, red foxes (Vulpes vulpes) or blue foxes (V. lagopus); the parasite must be consumed by these to complete its life cycle [4, 28]. Domestic dogs, wild dogs and red foxes (Vulpes vulpes) are also very common in the region covered by the present study, and these may also serve as definitive hosts; foxes, in particular, have unlimited access to farm animals, unlike dogs. Of the 13 wild carnivorous species known to carry unnamed Sarcocystis species [4] four of them, viz. raccoons (Procyon lotor), European badgers (Meles meles), otters (Lutra lutra) and mink (Mustella vision), are common in the area where the study was carried out.
The definitive hosts of S. morae and S. linearis are unclear. However, phylogenetic data indicate canids as most likely definitive host of these two species [18, 28]. It is possible that some Sarcocystis species in red deer may use other cervids as their principal intermediate hosts, and this may also indicate that many species of Sarcocystis are not as specific to their intermediate host as once believed.
Conclusion
Our study confirms the presence of Sarcocystis spp. in farmed fallow deer and is the first to identify three species of the genus Sarcocystis in them. Additionally, our results indicate that a wide range of mammals in different areas may harbour Sarcocystis species.
References
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2015) Sarcocystosis of animals and humans, 2nd edn. CRC Press Inc, Boca Raton
Tropilo J, Wisniewski J (2001) Sarcocystis spp. infection in free-living animals: wild boar (Sus scrofa L.), deer (Cervus elaphus L.), roe deer (Capreolus capreolus L.). Pol J Vet Sci 04:15–18
Gjerde B (2014) Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. https://doi.org/10.1017/S0031182013001819
Dubey J, Speer C, Fayer R (1989) Sarcocystosis of animals and man. CRC Press Inc, Boca Raton
Bittencourt MV, Meneses IDS, Ribeiro-Andrade M, de Jesus RF, de Araújo FR, Gondim LFP (2016) Sarcocystis spp. in sheep and goats: frequency of infection and species identification by morphological, ultrastructural, and molecular tests in Bahia, Brazil. Parasitol Res 115:1683–1689. https://doi.org/10.1007/s00436-016-4909-5
Morsy K, Abdel-Ghaffar F, Bin Dajem S, Abdel-Gaber R, El Gazar F (2018) First molecular characterization and morphological aspects of Sarcocystis fusiformis infecting water buffalo Bubalus bubalis in Egypt. Acta Parasitol 63:333–345. https://doi.org/10.1515/ap-2018-0038
Reissig EC, Moré G, Massone A, Uzal FA (2016) Sarcocystosis in wild red deer (Cervus elaphus) in Patagonia, Argentina. Parasitol Res 115:1773–1778. https://doi.org/10.1007/s00436-016-4915-7
Gjerde B, Luzón M, Alunda JM, de la Fuente C (2017) Morphological and molecular characteristics of six Sarcocystis spp. from red deer (Cervus elaphus) in Spain, including Sarcocystis cervicanis and three new species. Parasitol Res 116:2795–2811. https://doi.org/10.1007/s00436-017-5590-z
Stephan R, Loretz M, Eggenberger E, Grest P, Basso W, Grimm F (2012) Erster Nachweis von Sarcocystis hjorti bei der Fleischuntersuchung von Rothirschen in der Schweiz. Schweiz Arch Tierheilkd 154:539–542. https://doi.org/10.1024/0036-7281/a000403
Ota T, Nakano Y, Mizuno T, Shiozaki A, Hori Y, Yamanishi K, Hayakawa K, Hayakawa T, Fujimoto T, Nakamoto C, Maejima K, Wada Y, Terasoma F, Ohnishi T (2019) First case report of possible sarcocystis truncata-induced food poisoning in venison. Intern Med 58:2727–2730. https://doi.org/10.2169/internalmedicine.2817-19
Harris VC, van Vugt M, Aronica E, de Bree GJ, Stijnis C, Goorhuis A, Grobusch MP (2015) Human extraintestinal sarcocystosis: what we know, and what we don’t know. Curr Infect Dis Rep 17:42. https://doi.org/10.1007/s11908-015-0495-4
Fayer R, Esposito DH, Dubey JP (2015) Human infections with Sarcocystis species. Clin Microbiol Rev 28:295–311. https://doi.org/10.1128/CMR.00113-14
Entzeroth R, Chobotar B, Scholtyseck E, Neméseri L (1985) Light and electron microscope study of Sarcocystis sp. from the fallow deer (Cervus dama). Zeitschrift für Parasitenkd Parasitol Res 71:33–39. https://doi.org/10.1007/BF00932916
Poli A, Mancianti F, Marconcini A, Nigro M, Colagreco R (1988) Prevalence, ultrastructure of the cyst wall and infectivity for the dog and cat of Sarcocystis sp. from fallow deer (Cervus dama). J Wildl Dis 24:97–104. https://doi.org/10.7589/0090-3558-24.1.97
Hernández-Rodríguez S, Acosta I, Navarrete I (1992) Sarcocystisjorrini sp. nov. from the fallow deer Cervus dama. Parasitol Res 78:557–562. https://doi.org/10.1007/BF00936452
Wesemeier HH, Sedlaczek J (1995) One known Sarcocystis species and one found for the first time in fallow deer (Dama dama). Appl Parasitol 36:299–302
Rehbein S, Visser M, Jekel I, Silaghi C (2014) Endoparasites of the fallow deer (Dama dama) of the Antheringer Au in Salzburg, Austria. Wien Klin Wochenschr 126:S37–S41. https://doi.org/10.1007/s00508-014-0506-8
Lobão-Tello ER, Paredes E, Navarrete-Talloni MJ (2017) Sarcocystis spp. in red deer (Cervus elaphus), fallow deer (Dama dama), and pudu (Pudu pudu) in southern Chile. Pesqui Vet Bras 37:874–876. https://doi.org/10.1590/s0100-736x2017000800016
de Las Cuevas GED, Prakas P, Strazdaitė-Žielienė Ž, Martínez-González M, Rudaitytė-Lukošienė E, Butkauskas D, Servienė E, Habela MA, Calero-Bernal R (2019) Sarcocystis morae (Apicomplexa) in fallow deer (Dama dama) from Spain: ultrastructure and new host record. J Parasitol 105:813–815
Moré G, Schares S, Maksimov A, Conraths FJ, Venturini MC, Schares G (2013) Development of a multiplex real time PCR to differentiate Sarcocystis spp. affecting cattle. Vet Parasitol 197:85–94. https://doi.org/10.1016/j.vetpar.2013.04.024
Dahlgren SS, Gjerde B, Skirnisson K, Gudmundsdottir B (2007) Morphological and molecular identification of three species of Sarcocystis in reindeer (Rangifer tarandus tarandus) in Iceland. Vet Parasitol 149:191–198. https://doi.org/10.1016/j.vetpar.2007.08.015
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kolenda R, Ugorski M, Bednarski M (2014) Molecular characterization of Sarcocystis species from Polish roe deer based on ssu rRNA and cox1 sequence analysis. Parasitol Res 113:3029–3039. https://doi.org/10.1007/s00436-014-3966-x
Kolenda R, Schierack P, Zieba F, Zwijacz-Kozica T, Bednarski M (2015) First molecular characterization of Sarcocystis tenella in Tatra chamois (Rupicapra rupicapra tatrica) in Poland. Parasitol Res 114:3885–3892. https://doi.org/10.1007/s00436-015-4619-4
Pyziel AM, Demiaszkiewicz AW (2009) Sarcocystis cruzi (Protozoa: Apicomplexa: Sarcocystiidae) infection in European bison (Bison bonasus) from Białowieza Forest, Poland. Wiad Parazytol 55:31–34
Wesemeier HH, Sedlaczek J (1995) One known Sarcocystis species and two found for the first time in red deer and wapiti (Cervus elaphus) in Europe. Appl Parasitol 36:245–251
Gjerde B, Giacomelli S, Bianchi A, Bertoletti I, Mondani H, Gibelli LR (2017) Morphological and molecular characterization of four Sarcocystis spp., including Sarcocystis linearis n. sp., from roe deer (Capreolus capreolus) in Italy. Parasitol Res 116:1317–1338. https://doi.org/10.1007/s00436-017-5410-5
Dahlgren SS, Gjerde B (2007) Genetic characterisation of six Sarcocystis species from reindeer (Rangifer tarandus tarandus) in Norway based on the small subunit rRNA gene. Vet Parasitol 146:204–213. https://doi.org/10.1016/j.vetpar.2007.02.023
Dahlgren SS, Gjerde B (2009) Sarcocystis in Norwegian roe deer (Capreolus capreolus): Molecular and morphological identification of Sarcocystis oviformis n. sp. and Sarcocystis gracilis and their phylogenetic relationship with other Sarcocystis species. Parasitol Res 104:993–1003. https://doi.org/10.1007/s00436-008-1281-0
Januškevičius V, Januškevičiene G, Paulauskas V, Petkevičius S (2016) Prevalence and intensity of sarcosporidia infection in wild cloven-hoofed hunted for food in Lithuania. Vet ir Zootech 73:27–31
Malakauskas M, Grikienienė J (2002) Sarcocystis infection in wild ungulates in Lithuania. Acta Zool Litu 12:372–380. https://doi.org/10.1080/13921657.2002.10512527
Funding
This study was funded by W. Stefański Institute of Parasitology Polish Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
WC and BM devised the study concept and conducted part of the lab work; WC, BM, JB analyzed the results and wrote the first draft. JB and KG, KB carried out the molecular study and generated the sequences, designed the figures and edited the manuscript. KB was involved in the construction of the phylogenetic tree. ŻSB and MB collected the samples. All authors have agreed with the content of the manuscript. The authors declare no conflict of interests.
Ethical approval
This article is not under consideration or published elsewhere; no material in the paper has appeared or will appear on a preprint server. All animals belong to the Witold Stefański Institute of Parasitology, Polish Academy of Sciences. The Breeding Station is located in Kosewo Górne. The experimental protocol was approved by Scientific Board of the Witold Stefański Institute of Parasitology PAS.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cabaj, W., Bień-Kalinowska, J., Goździk, K. et al. Molecular identification of sarcocysts from tissue of fallow deer (Dama dama) farmed in the open pasture system based on ssu rRNA gene. Acta Parasit. 65, 354–360 (2020). https://doi.org/10.2478/s11686-019-00159-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11686-019-00159-0