Abstract
Purpose
Haemoparasitic diseases are among the important factors that threaten cattle health and productivity especially in the sub-Saharan region. In Nigeria, their detection using sensitive molecular techniques is scanty. This study was designed to investigate and to reevaluate the repertoire of haemoparasites of cattle in Ibadan, Nigeria with a comparative evaluation of light microscopy (LM) and polymerase chain reaction (PCR) methods.
Methods
Blood samples from 100 cattle slaughtered at Ibadan abattoirs were examined using LM and PCR techniques for haemoparasite detection. The PCR reactions using three primer sets targeting the 16S rRNA genes for Hemoplasma spp. and Anaplasma/Ehrlichia spp. and 18S rRNA genes of Babesia/Theleiria spp. were done. A few randomly selected amplicons from each set were sequenced and analysed.
Results
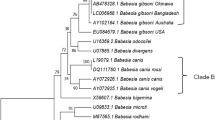
A total infection rate of 34% by LM including Hemoplasma spp. (17%), Anaplasma spp. (16%), microfilaria (5%) and Trypanosoma spp. (12%) was recorded. While, 86% positivity was recorded with PCR amplification as follows: Hemoplasma spp. (64%), Babesia/Theleiria spp. (46%) and Anaplasma/Ehrlichia spp. (5%). Comparison of LM and PCR findings showed that no LM Anaplasma spp.-positive samples and 7 out of the 17 LM hemoplasma-positive cattle were confirmed by PCR. In addition, LM led to misdiagnosis of 46 Babesia/Theleiria spp.-positive samples. Amplicon sequencing and phylogenetic analysis of Babesia/Theileria spp.-positive samples revealed Theileria velifera and Theileria annulata. In the Anaplasma/Ehrlichia spp.-positive samples, only Anaplasma marginale was characterized. Mycoplasma wenyonii, “Candidatus Mycoplasma haemobos” and Pseudomonas fluorescens like were characterized among the hemoplasma-infected cattle.
Conclusions
The first report of “Candidatus Mycoplasma haemobos” and Pseudomonas fluorescens like in Nigerian cattle is herewith documented. The alarming LM misdiagnosis of haemoparasites during this study confirms its limitations as it fails to identify many parasites and emphasizes the need for inclusion of molecular techniques to improve their detection. The study also shows for the first time the high prevalence of haemotropic mycoplasma in Nigerian cattle via molecular diagnostic methods, thus indicating a strong need for the investigation of their zoonotic implications.



Similar content being viewed by others
References
Bock R, Jackson L, De Vos A, Jorgensen W (2004) Babesiosis of cattle. Parasitol 129:S247–S269
EFSA (European Food Safety Authority) (2010) Scientific opinion on geographic distribution of tick-borne infections and their Vectors in Europe and the other Regions of the Mediterranean Basin. EFSA J 8(9):1723
Ellis JT, Morrison DA, Reichel MP (2003) Genomics and its impact on parasitology and the potential for development of new parasite control methods. DNA Cell Biol 32:395–403
CFSPH (2008) Bovine babesiosis. The Centre for Food Security and Public Health, Tick Fever, Cattle Fever, Texas Fever, Piroplasmosis, Redwater. Iowa State University, Ames, Iowa 50011. http://www.cfsph.iastate.edu/Factsheets/pdfs/bovine_babesiosis.pdf
Akande FA, Takeet MI, Makanju OA (2010) Haemoparasites of cattle in Abeokuta, South West Nigeria. Sci World J 5:19–21
Kamani J, Sannusi A, Egwu OK, Dogo GI, Tanko TJ, Kemza S, Tafarki AE, Gbise DS (2010) Prevalence and significance of haemoparasitic infections of cattle in North-Central, Nigeria. Vet World 3(10):445–448
Okorafor UP, Nzeako SO (2014) Prevalence of haemoparasites of cattle from three abattoirs in Ibadan Metropolis, Nigeria. Int J Sci Res Environ Sci 2(7):244–249
Maria R, Jeffrey AG (1996) Babesiosis. Clin Infect Dis 22:611–615
Sasaki M, Omobowale O, Tozuka M, Ohta K, Matsuu A, Nottidge HO, Hirata H, Ikadai H, Oyamada T (2007) Molecular survey of Babesia canis in dogs in Nigeria. J Vet Med Sci 69(11):1191–1193
Kamani J, Baneth G, Mumcuoglu KY, Waziri NE, Eyal O, Guthmann Y, Harrus S (2013) Molecular detection and characterization of tick-borne pathogens in dogs and ticks from Nigeria. PLoS Negl Trop Dis 7(3):2108
Adamu M, Troskie M, Oshadu DO, Malatji DP, Penzhorn BL, Matjila PT (2014) Occurrence of tick-transmitted pathogens in dogs in Jos, Plateau State, Nigeria. Parasit Vect 7(1):119
Happi AN, Toepp AJ, Ugwu CA, Petersen CA, Sykes JE (2018) Detection and identification of blood-borne infections in dogs in Nigeria using light microscopy and the polymerase chain reaction. Vet Parasitol Reg Stud Rep 11:55–60
Swallow BM (2000) Impacts of trypanosomosis on African agriculture. PAAT technical and Scientific Series, vol 2. FAO, Rome
Leeflang P, Ilemobade AA (1977) Tick-borne diseases of domestic animals in Northern Nigeria. Trop Anim Health Pro 9(4):211–218
Obi TU, Anosa VO (1980) Haematological studies on domestic animals in Nigeria: IV. Clinico-haematological features of bovine trypanosomiasis, theileriosis, anaplasmosis, eperythrozoonosis and helminthiasis. J Vet Med 27(9–10):789–797
Lorusso V, Wijnveld M, Majekodunmi AO, Dongkum C, Fajinmi A, Dogo AG, Thrusfield M, Mugenyi A, Vaumourin E, Igweh AC, Jongejan F (2016) Tick-borne pathogens of zoonotic and veterinary importance in Nigerian cattle. Parasit Vect 9(1):217
Maggi RG, Compton SM, Trull CL, Mascarelli PE, Mozayeni BR, Breitschwerdt EB (2013) Infection with hemotropic Mycoplasma species in patients with or without extensive arthropod or animal contact. J Clin Microbiol 51(10):3237–3241
McAuliffe L, Lawes J, Bell S, Barlow A, Aylin R, Nicholas R (2006) The detection of mycoplasma (formerly Eperythrozoon) wenyonii by 16S rDNA PCR and denaturing gradient gel electrophoresis. Vet Microbiol 117:292–296
Tagawa M, Matsumoto K, Inokuma H (2008) Molecular detection of Mycoplasma wenyonii and ‘Candidatus Mycoplasma haemobos’ in cattle in Hokkaido, Japan. Vet Microbiol 132:177–180
Meli ML, Willi B, Dreher UM, Cattori V, Knubben-Schweizer G, Nuss K, Braun U, Lutz H, Hofmann-Lehmann R (2010) Identification, molecular characterization, and occurrence of two bovine Hemoplasma spp. in Swiss cattle and development of real-time TaqMan quantitative PCR assays for diagnosis of bovine hemoplasma infections. J Clin Microbiol 48(10):3563–3568
Uilenberg G (2009) Candidatus Mycoplasma haemobos. Vet Microbiol 138(1–2):200–201
Girotto A, Zangirólamo AF, Bogado ALG, Souza ASL, da Silva GCF, Garcia JL, Boas AAV, Biondo AW, Vidotto O (2012) Molecular detection and occurrence of ‘Candidatus Mycoplasma haemobos’ in dairy cattle of Southern Brazil. Rev Bras Parasitol Vet 21(3):342–344
Picozzi K, Tilley A, Fèvre EM, Coleman PG, Magona JW, Odiit M, Eisler MC, Welburn SC (2002) The diagnosis of trypanosome infections: applications of novel technology for reducing disease risk. Afr J Biotechnol 1(2):39–45
Latimer KS (2011) Duncan and Prasse’s veterinary laboratory medicine: clinical pathology, 5th edn. Wiley-Blackwell, Oxford
Schouls LM, Van De Pol I, Rijpkema SGT, Schot CS (1999) Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella spp. in Dutch Ixodes ricinus ticks. J Clin Microbiol 37:2215–2222
Bekker CPJ, de Vos S, Taoufik A, Sparagano OAE, Jongejan F (2002) Simultaneous detection of Anaplasma and Ehrlichia spp. in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridisation. Vet Microbiol 89:223–238
Georges K, Loria GR, Riili S, Greco A, Caracappa S, Jongejan F, Sparagano O (2001) Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet Parasitol 99:273–286
Fujihara Y, Sasaoka F, Suzuki J, Watanabe Y, Fujihara M, Ooshita K, Ano H, Harasawa R (2011) Prevalence of hemoplasma infection among cattle in the western part of Japan. J Vet Med Sci 73(12):1653–1655
Nishizawa I, Sato M, Fujihara M, Sato S, Harasawa R (2010) Differential detection of hemotropic Mycoplasma spp. in cattle by melting curve analysis of PCR products. J Vet Med Sci 72(1):77–79
Su QL, Song HQ, Lin RQ, Yuan ZG, Yang JF, Zhao GH, Huang WY, Zhu XQ (2010) The detection of “Candidatus Mycoplasma haemobos” in cattle and buffalo in China. Trop Anim Health Pro 42(8):1805–1808
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10(3):512–526
Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, Ceci L, Buonavoglia C (2007) Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet Microbiol 124(1–2):107–114
Hoelzle LE (2008) Haemotrophic mycoplasmas: recent advances in Mycoplasma suis. Vet Microbiol 130:215–226
Aylin RD, Bisgaard-Frantzen S, Adler A, Blowey RW, Barlow AM, Millar MF, van der Burgt GM (2012) Detection of ‘Candidatus Mycoplasma haemobos’, Mycoplasma wenyonii and Anaplasma phagocytophilum from cattle in England. Vet Rec 170(21):543a
McFadden AMJ, Ha HJ, Donald JJ, Bueno IM, van Andel M, Thompson JC, Tisdall JC, Pulford DJ (2016) Investigation of bovine hemoplasmas and their association with anaemia in New Zealand cattle. N Z Vet J 64:65–68
Olabode HOK, Jegede OC, Ajagbonna OP, Adah BMJ, Obafemi FA (2014) Evaluation of haemoparasites in trade cattle slaughtered in Jos Abattoir, Plateau State-Nigeria. Int J Livest Res 4(1):113–120
Birkenheuer AJ, Breitschwerdt EB, Alleman AR, Pitulle C (2002) Differentiation of Haemobartonella canis and Mycoplasma haemofelis on the basis of comparative analysis of gene sequence. Am J Vet Res 63(10):1385–1388
Criado-Fornelio A, Martinez-Marcos A, Buling-Saraña A, Barba-Carretero JC (2003) Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet Microbiol 93(4):307–317
Willi B, Filoni C, Catão-Dias JL, Cattori V, Meli ML, Vargas A, Martínez F, Roelke ME, Ryser-Degiorgis MP, Leutenegger CM, Lutz H, Hofmann-Lehmann R (2007) Worldwide occurrence of feline hemoplasma infections in wild felid species. J Clin Microbiol 45:1159–1166
Bauer N, Balzer HJ, Thüre S, Moritz A (2008) Prevalence of feline haemotropic mycoplasmas in convenience samples of cats in Germany. J Feline Med Surg 10:252–258
Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB (2013) Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasit Vect 6(1):103
Yuan CL, Liang AB, Yao CB, Yang ZB, Zhu JG, Cui L, Yu F, Zhu NY, Yang XW, Hua XG (2009) Prevalence of Mycoplasma suis (Eperythrozoon suis) infection in swine and swine-farm workers in Shanghai, China. Am J Vet Res 70:890–894
Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB (2010) Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J Clin Microbiol 48:3782–3785
Steer JA, Tasker S, Barker EN, Jensen J, Mitchell J, Stocki T, Chalker VJ, Hamon M (2011) A novel hemotropic Mycoplasma (hemoplasma) in a patient with hemolytic anemia and pyrexia. Clin Infect Dis 53:e147–e151
Bihonegn W, Haimanot D, Tadele K, Tilahun Z, Girma K (2015) Study on the prevalence of bovine babesiosis and its associated risk factors in and around Assosa Woreda, Benishangul Gumuz Regional State, Western Ethiopia. Researcher 7(8):33–39
Saad F, Khan K, Ali S, ul Akbar N (2015) Zoonotic significance and prophylactic measure against babesiosis. Int J Curr Microbiol App Sci 4(7):938–953
Ademola IO, Onyechi TE (2013) Haemoparasites and haematological parameters of slaughtered ruminants and pigs at Bodija Abattoir, Ibadan, Nigeria. Afr J Biomed Res 16:101–105
Tagawa M, Ybañez AP, Matsumoto K, Yokoyama N, Inokuma H (2012) Prevalence and risk factor analysis of bovine hemoplasma infection by direct PCR in eastern Hokkaido, Japan. J Vet Med Sci 74:1171–1176
Adejinmi JO, Sadiq NA, Fashanu SO, Lasisi OT, Ekundayo S (2004) Study on the blood parasite of sheep in Ibadan, Nigeria. Afr J Biomed Res 7:42–43
Tagawa M, Matsumoto K, Yokoyama N, Inokuma H (2010) Comparison of the effect of two Hemoplasma spp. on hematological parameters in cattle. J Vet Med Sci 72(1):113–115
Salih DA, El Hussein AM, Hayat M, Taha KM (2003) Survey of Theileria lestoquardi antibodies among Sudanese sheep. Vet Parasitol 111(4):361–367
Al-Hamidhi S, Weir W, Kinnaird J, Tageledin M, Beja-Pereira A, Morrison I, Thompson J, Tait A, Shiels B, Babiker HA, Babiker HA (2016) Theileria lestoquardi displays reduced genetic diversity relative to sympatric Theileria annulata in Oman. Infect Genet Evol 43:297–306
Ali AM, Salih DA, Njahira MN, Hassan SK, El Hussein AM, Liu Z, Yin H, Pelle R, Skilton RA (2017) Genotyping of Theileria lestoquardi from sheep and goats in Sudan to support control of Malignant Ovine Theileriosis. Vet Parasitol 239:7–14
Awad H, Al-Hamidhi S, El Hussein ARM, Yousif YM, Taha KM, Salih DA, Weir W, Babiker HA (2018) Theileria lestoquardi in Sudan is highly diverse and genetically distinct from that in Oman. Infect Genet Evol 62:46–52
Leemans I, Brown D, Hooshmand-Rad P, Kirvar E, Uggla A (1999) Infectivity and cross-immunity studies of Theileria lestoquardi and Theileria annulata in sheep and cattle: in vivo responses. Vet Parasitol 82(3):179–192
Peters IR, Helps CR, McAuliffe L, Neimark H, Lappin MR, Gruffydd-Jones TJ, Day MJ, Hoelzle LE, Willi B, Meli M, Hofmann-Lehmann R (2008) RNase P RNA gene (rnpB) phylogeny of Hemoplasmas and other Mycoplasma spp. J Clin Microbiol 46(5):1873–1877
Watanabe Y, Fujihara M, Obara H, Matsubara K, Yamauchi K, Harasawa R (2010) Novel hemoplasma spp detected in free-ranging sika deer (Cervus nippon). J Vet Med Sci 72(11):1527–1530
Ade J, Niethammer F, Schade B, Schilling T, Hoelzle K, Hoelzle LE (2018) Quantitative analysis of Mycoplasma wenyonii and “Candidatus Mycoplasma haemobos” infections in cattle using novel gapN-based realtime PCR assays. Vet Microbiol 220:1–6
Hoelzle K et al (2011) Detection of ‘Candidatus Mycoplasma haemobos’ in cattle with anaemia. Vet J 187:408–410
Kwon SW, Kim JS, Park IC, Yoon SH, Park DH, Lim CK, Go SJ (2003) Pseudomonas koreensis sp. nov., Pseudomonas umsongensis sp. nov. and Pseudomonas jinjuensis sp. nov., novel spp from farm soils in Korea. Int J Syst Evol Microbiol 53(1):21–27
Ganeshan G, Kumar AM (2005) Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. J Plant Interact 1(3):123–134
Lin H, Hu S, Liu R, Chen P, Ge C, Zhu B, Guo L (2016) Genome sequence of Pseudomonas koreensis CRS05-R5, an antagonistic bacterium isolated from rice paddy field. Front Microbiol 7:1756
Chiesa F, Lomonaco S, Nucera D, Garoglio D, Dalmasso A, Civera T (2014) Distribution of Pseudomonas spp. in a dairy plant affected by occasional blue discoloration. Int J Food Saf 3:245–248
Cho ST, Chang HH, Egamberdieva D, Kamilova F, Lugtenberg B, Kuo CH (2015) Genome analysis of Pseudomonas fluorescens PCL1751: a rhizobacterium that controls root diseases and alleviates salt stress for its plant host. PLoS One 10(14):231
Rafikova GF, Korshunova TY, Minnebaev LF, Chetverikov SP, Loginov ON (2016) A new bacterial strain, Pseudomonas koreensis IB-4, as a promising agent for plant pathogen biological control. Microbiol 85(3):333–341
Schlusselhuber M, Godard J, Sebban M, Bernay B, Garon D, Seguin V, Oulyadi H, Desmasures N (2018) Characterization of Milkisin, a novel lipopeptide with antimicrobial properties produced by Pseudomonas sp. UCMA 17988 isolated from bovine raw milk. Front Microbiol 9:1030
Ming L, Yi L, Hasi S, He J, Hai L, Wang Z, Gu F, Qiao X (2017) Comparative analysis of fecal microbial communities in cattle and Bactrian camels. PLoS One 12(3):e0173062
Scaccabarozzi L, Leoni L, Ballarini A, Barberio A, Locatelli C, Casula A, Bronzo V, Pisoni G, Jousson O, Morandi S, Rapetti L (2015) Pseudomonas aeruginosa in dairy goats: genotypic and phenotypic comparison of intramammary and environmental isolates. PLoS One 10(11):e0142973
Serrano I, De Vos D, Santos JP, Bilocq F, Leitão A, Tavares L, Pirnay JP, Oliveira M (2016) Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure. BMC Vet Res 13(1):58
Acknowledgements
The authors are gratefully indebted to Professor Christian Happi for his unceasing support and for allowing us to use his laboratory facilities at the African Center of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Ede, Nigeria, for the molecular analyses carried out in this study. Our unreserved thanks and gratitude goes particularly to Mrs. Philonema Eromon and Mr. Adeyemi T Kayode for their technical assistance in the molecular laboratory at the ACEGID. We immensely appreciate Mrs Ademakinwa Josephine and Mrs Ayomikun Adeyeyi for their support in the haematology work at the Clinical Pathology Laboratory, Department of Veterinary Pathology, University of Ibadan, Nigeria.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Statement of animal rights
The present study was carried out by collecting blood from cattle at the slaughtering point during bleeding. This was done with the consent of the owners of the respective cattle at the local municipal abattoir in Ibadan, Nigeria. Therefore as we were not on our own handling the animals, we thought there was no need for ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Happi, A.N., Osifade, O., Oluniyi, P.E. et al. Comparison of Light Microscopy and Polymerase Chain Reaction for the Detection of Haemoparasites in Cattle in Nigeria. Acta Parasit. 65, 44–56 (2020). https://doi.org/10.2478/s11686-019-00123-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11686-019-00123-y