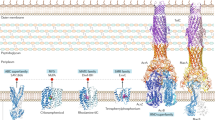
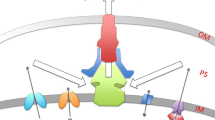
Abstract
Drug resistance in bacteria, and especially resistance to multiple antibacterials, has attracted much attention in recent years. In addition to the well known mechanisms, such as inactivation of drugs and alteration of targets, active efflux is now known to play a major role in the resistance of many species to antibacterials. Drug-specific efflux (e.g. that of tetracycline) has been recognised as the major mechanism of resistance to this drug in Gram-negative bacteria. In addition, we now recognise that multidrug efflux pumps are becoming increasingly important. Such pumps play major roles in the antiseptic resistance of Staphylococcus aureus, and fluoroquinolone resistance of S. aureus and Streptococcus pneumoniae. Multidrug pumps, often with very wide substrate specificity, are not only essential for the intrinsic resistance of many Gram-negative bacteria but also produce elevated levels of resistance when overexpressed. Paradoxically, ‘advanced’ agents for which resistance is unlikely to be caused by traditional mechanisms, such as fluoroquinolones and β-lactams of the latest generations, are likely to select for overproduction mutants of these pumps and make the bacteria resistant in one step to practically all classes of antibacterial agents. Such overproduction mutants are also selected for by the use of antiseptics and biocides, increasingly incorporated into consumer products, and this is also of major concern. We can consider efflux pumps as potentially effective antibacterial targets. Inhibition of efflux pumps by an efflux pump inhibitor would restore the activity of an agent subject to efflux. An alternative approach is to develop antibacterials that would bypass the action of efflux pumps.






Similar content being viewed by others
References
Levy SB. Antibiotic resistance: an ecological imbalance. Ciba Found Symp 1997; 207: 1–9
Normark BH, Normark S. Evolution and spread of antibiotic resistance. J Intern Med 2002 Aug; 252(2): 91–106
Ball PR, Chopra I, Eccles SJ. Accumulation of tetracyclines by Escherichia coli K-12. Biochem Biophys Res Commun 1977 Aug 22; 77(4): 1500–7
Ball PR, Shales SW, Chopra I. Plasmid-mediated tetracycline resistance in Escherichia coli involves increased efflux of the antibiotic. Biochem Biophys Res Commun 1980 Mar 13; 93(1): 74–81
Levy SB, McMurry L. Plasmid-determined tetracycline resistance involves new transport systems for tetracycline. Nature 1978 Nov 2; 276(5683): 90–2
McMurry L, Petrucci Jr RE, Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A 1980 Jul; 77(7): 3974–7
Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta 1976 Nov 11; 455(1): 152–62
Institute of Genome Research (TIGR) microbial database [online]. Available from URL: http://www.tigr.org/tdb/mdb/mdbcomplete.html [Accessed 2003 Nov 11]
Lomovskaya O, Warren MS, Lee V. Efflux mechanisms: molecular and clinical aspects. In: Hughes D, Andersson DI, editors. Antibiotic development and resistance. London: Taylor and Francis 2001: 65–90
Poole K. Outer membranes and efflux: the path to multidrug resistance in Gram-negative bacteria. Curr Pharm Biotechnol 2002 Jun; 3(2): 77–98
Paulsen IT, Lewis K. Microbial multidrug efflux. Wynmondham: Horizon Press, 2002
Fath MJ, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev 1993 Dec; 57(4): 995–1017
Higgins CF. ABC transporters: physiology, structure and mechanism: an overview. Res Microbiol 2001 Apr–May; 152(3–4): 205–10
Pao SS, Paulsen IT, Saier Jr MH. Major facilitator superfamily. Microbiol Mol Biol Rev 1998 Mar; 62(1): 1–34
Brown MH, Paulsen IT, Skurray RA. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol 1999 Jan; 31(1): 394–5
Paulsen IT, Skurray RA, Tam R, et al. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol 1996 Mar; 19(6): 1167–75
Saier Jr MH, Tam R, Reizer A, et al. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol 1994 Mar; 11(5): 841–7
Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 1996 Oct; 178(20): 5853–9
Paulsen IT, Park JH, Choi PS, et al. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett 1997 Nov 1; 156(1): 1–8
Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol 1999 Apr 2; 287(3): 695–715
Dinh T, Paulsen IT, Saier Jr MH. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol 1994 Jul; 176(13): 3825–31
Saier Jr MH, Paulsen IT, Sliwinski MK, et al. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J 1998 Mar; 12(3): 265–74
Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol 1992; 8: 67–113
Bolhuis H, van Veen HW, Brands JR, et al. Energetics and mechanism of drug transport mediated by the lactococcal multidrug transporter LmrP. J Biol Chem 1996 Sep 27; 271(39): 24123–8
Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol 2001 Oct; 183(19): 5639–44
Marger MD, Saier Jr MH. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci 1993 Jan; 18(1): 13–20
Saier Jr MH, Beatty JT, Goffeau A, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol 1999 Nov; 1(2): 257–79
Yoshida H, Bogaki M, Nakamura S, et al. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol 1990 Dec; 172(12): 6942–9
Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A 1992 Oct 1; 89(19): 8938–42
Morita Y, Kodama K, Shiota S, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 1998 Jul; 42(7): 1778–82
Chung YJ, Saier Jr MH. SMR-type multidrug resistance pumps. Curr Opin Drug Discov Devel 2001 Mar; 4(2): 237–45
Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev 1996 Dec; 60(4): 575–608
Grinius L, Dreguniene G, Goldberg EB, et al. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 1992 Mar; 27(2): 119–29
Schuldiner S, Lebendiker M, Yerushalmi H. EmrE, the smallest ion-coupled transporter, provides a unique paradigm for structure-function studies. J Exp Biol 1997 Jan; 200 (Pt 2): 335–41
Tseng TT, Gratwick KS, Kollman J, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol 1999 Aug; 1(1): 107–25
Grosse C, Grass G, Anton A, et al. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J Bacteriol 1999 Apr; 181(8): 2385–93
Droge M, Puhler A, Selbitschka W. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol Gen Genet 2000 Apr; 263(3): 471–82
Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis 1998 Aug; 27 Suppl. 1: S32–41
Ma D, Cook DN, Alberti M, et al. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol 1993 Oct; 175(19): 6299–313
Fralick JA. Evidence that tolC is required for functioning of the mar/acrAB efflux pump of Escherichia coli. J Bacteriol 1996 Oct; 178(19): 5803–5
Poole K, Krebes K, McNally C, et al. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol 1993 Nov; 175(22): 7363–72
Li XZ, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1995 Sep; 39(9): 1948–53
Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother 1989 Nov; 33(11): 1831–6
Li XZ, Livermore DM, Nikaido H. Role of efflux pump (s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother 1994 Aug; 38(8): 1732–41
Li XZ, Ma D, Livermore DM, et al. Role of efflux pump (s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob Agents Chemother 1994 Aug; 38(8): 1742–52
Nikaido H. The role of outer membrane and efflux pumps in the resistance of gram-negative bacteria: can we improve drug access? Drug Resist Updat 1998; 1: 93–8
Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol 2001 Oct; 183(20): 5803–12
Nakamura H. Gene-controlled resistance to acriflavine and other basic dyes in Escherichia coli. J Bacteriol 1965 Jul; 90(1): 8–14
Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci U S A 1999 Jun 22; 96(13): 7190–5
Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM 4454. Antimicrob Agents Chemother 2001 Dec; 45(12): 3375–80
Palumbo JD, Kado CI, Phillips DA. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol 1998 Jun; 180(12): 3107–13
Peng WT, Nester EW. Characterization of a putative RND-type efflux system in Agrobacterium tumefaciens. Gene 2001 May 30; 270(1–2): 245–52
Krummenacher P, Narberhaus F. Two genes encoding a putative multidrug efflux pump of the RND/MFP family are cotranscribed with an rpoH gene in Bradyrhizobium japonicum. Gene 2000 Jan 11; 241(2): 247–54
Burns JL, Wadsworth CD, Barry JJ, et al. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob Agents Chemother 1996 Feb; 40(2): 307–13
Moore RA, DeShazer D, Reckseidler S, et al. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother 1999 Mar; 43(3): 465–70
Lin J, Michel LO, Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 2002 Jul; 46(7): 2124–31
Pradel E, Pages JM. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob Agents Chemother 2002 Aug; 46(8): 2640–3
Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol 2000 Mar; 182(6): 1754–6
Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J Bacteriol 2002 Dec; 184(23): 6490–8
Ma D, Cook DN, Hearst JE, et al. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol 1994 Dec; 2(12): 489–93
Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J Bacteriol 2002 Aug; 184(15): 4168–76
Nagakubo S, Nishino K, Hirata T, et al. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol 2002 Aug; 184(15): 4161–7
Nishino K, Yamaguchi A. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J Bacteriol 2002 Apr; 184(8): 2319–23
Sanchez L, Pan W, Vinas M, et al. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol 1997 Nov; 179(21): 6855–7
Hagman KE, Pan W, Spratt BG, et al. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 1995 Mar; 141 (Pt 3): 611–22
Lucas CE, Balthazar JT, Hagman KE, et al. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol 1997 Jul; 179(13): 4123–8
Lee EH, Shafer WM. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol Microbiol 1999 Aug; 33(4): 839–45
Ikeda T, Yoshimura F. A resistance-nodulation-cell division family xenobiotic efflux pump in an obligate anaerobe, Porphyromonas gingivalis. Antimicrob Agents Chemother 2002 Oct; 46(10): 3257–60
Poole K, Tetro K, Zhao Q, et al. Expression of the multidrug resistance operon MexA-MexB-OprM in Pseudomonas aeruginosa: MexR encodes a regulator of operon expression. Antimicrob Agents Chemother 1996 Sep; 40(9): 2021–8
Poole K, Gotoh N, Tsujimoto H, et al. Overexpression of the MexC-MexD-OprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 1996 Aug; 21(4): 713–24
Kohler T, Michea-Hamzehpour M, Henze U, et al. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol 1997 Jan; 23(2): 345–54
Aires JR, Kohler T, Nikaido H, et al. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 1999 Nov; 43(11): 2624–8
Mine T, Morita Y, Kataoka A, et al. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother 1999 Feb; 43(2): 415–7
Westbrock-Wadman S, Sherman DR, Hickey MJ, et al. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 1999 Dec; 43(12): 2975–83
Aendekerk S, Ghysels B, Cornelis P, et al. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 2002 Aug; 148 (Pt 8): 2371–81
Chuanchuen R, Narasaki CT, Schweizer HP. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J Bacteriol 2002 Sep; 184(18): 5036–44
Kieboom J, Dennis JJ, de Bont JA, et al. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem 1998 Jan 2; 273(1): 85–91
Ramos JL, Duque E, Godoy P, et al. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J Bacteriol 1998 Jul; 180(13): 3323–9
Fukumori F, Hirayama H, Takami H, et al. Isolation and transposon mutagenesis of a Pseudomonas putida KT2442 toluene-resistant variant: involvement of an efflux system in solvent resistance. Extremophiles 1998 Nov; 2(4): 395–400
Mosqueda G, Ramos JL. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J Bacteriol 2000 Feb; 182(4): 937–43
Rojas A, Duque E, Mosqueda G, et al. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J Bacteriol 2001 Jul; 183(13): 3967–73
Li XZ, Zhang L, Poole K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 2002 Feb; 46(2): 333–43
Zhang L, Li XZ, Poole K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 2001 Dec; 45(12): 3497–503
Alonso A, Martinez JL. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 2000 Nov; 44(11): 3079–86
Kumar A, Worobec EA. Fluoroquinolone resistance of Serratia marcescens: involvement of a proton gradient-dependent efflux pump. J Antimicrob Chemother 2002 Oct; 50(4): 593–6
Nikaido H, Basina M, Nguyen V, et al. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol 1998 Sep; 180(17): 4686–92
Lacroix FJ, Cloeckaert A, Grepinet O, et al. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol Lett 1996 Jan 15; 135(2–3): 161–7
Sulavik MC, Houseweart C, Cramer C, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother 2001 Apr; 45(4): 1126–36
Furukawa H, Tsay JT, Jackowski S, et al. resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol 1993 Jun; 175(12): 3723–9
Bohn C, Bouloc P. The Escherichia coli cmlA gene encodes the multidrug efflux pump Cmr/MdfA and is responsible for isopropyl-β-D-thiogalactopyranoside exclusion and spectinomycin sensitivity. J Bacteriol 1998 Nov; 180(22): 6072–5
Nilsen IW, Bakke I, Vader A, et al. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol 1996 Jun; 178(11): 3188–93
Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J 1999 Feb 15; 18(4): 822–32
Mine T, Morita Y, Kataoka A, et al. Evidence for chloramphenicol/H+ antiport in Cmr (MdfA) system of Escherichia coli and properties of the antiporter. J Biochem (Tokyo) 1998 Jul; 124(1): 187–93
Dorman CJ, Foster TJ, Shaw WV. Nucleotide sequence of the R26 chloramphenicol resistance determinant and identification of its gene product. Gene 1986; 41(2–3): 349–53
Ploy MC, Courvalin P, Lambert T. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob Agents Chemother 1998 Oct; 42(10): 2557–63
Toro CS, Lobos SR, Calderon I, et al. Clinical isolate of a porinless Salmonella typhi resistant to high levels of chloram-phenicol. Antimicrob Agents Chemother 1990 Sep; 34(9): 1715–9
Bissonnette L, Champetier S, Buisson JP, et al. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol 1991 Jul; 173(14): 4493–502
Kim E, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol 1996; 40(9): 665–9
Bolton LF, Kelley LC, Lee MD, et al. Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol 1999 May; 37(5): 1348–51
Boyd D, Peters GA, Cloeckaert A, et al. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 2001 Oct; 183(19): 5725–32
White DG, Hudson C, Maurer JJ, et al. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J Clin Microbiol 2000 Dec; 38(12): 4593–8
Schuldiner S, Granot D, Steiner S, et al. Precious things come in little packages. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 155–62
Yerushalmi H, Schuldiner S. A model for coupling of H+ and substrate fluxes based on ‘time-sharing’ of a common binding site. Biochemistry 2000 Dec 5; 39(48): 14711–9
Yerushalmi H, Schuldiner S. A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett 2000 Jun 30; 476(1–2): 93–7
Yerushalmi H, Schuldiner S. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem 2000 Feb 25; 275(8): 5264–9
Chung YJ, Saier Jr MH. Overexpression of the Escherichia colisugE gene confers resistance to a narrow range of quaternary ammonium compounds. J Bacteriol 2002 May; 184(9): 2543–5
Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother 1992 Apr; 36(4): 695–703
Thanassi DG, Suh GS, Nikaido H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol 1995 Feb; 177(4): 998–1007
Yamaguchi A, Udagawa T, Sawai T. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem 1990 Mar 25; 265(9): 4809–13
Linton KJ, Higgins CF. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol 1998 Apr; 28(1): 5–13
Allikmets R, Gerrard B, Court D, et al. Cloning and organization of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene 1993 Dec 22; 136(1–2): 231–6
Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 1994 Apr 15; 264(5157): 382–8
Poole K, Heinrichs DE, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol 1993 Nov; 10(3): 529–44
Nikaido H. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin Cell Dev Biol 2001 Jun; 12(3): 215–23
Poole K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 255–64
Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 2000 Aug 31; 406(6799): 959–64
Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1992 Sep; 36(9): 1847–51
Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1995 Mar; 39(3): 645–9
Srikumar R, Paul CJ, Poole K. Influence of mutations in the MexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 2000 Mar; 182(5): 1410–4
Ziha-Zarifi I, Llanes C, Köhler T, et al. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa over-expressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother 1999 Feb; 43(2): 287–91
Köhler T, Kok M, Michea-Hamzehpour M, et al. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1996 Oct; 40(10): 2288–90
Li XZ, Zhang L, Srikumar R, et al. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1998 Feb; 42(2): 399–403
Schweizer HP. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother 1998 Feb; 42(2): 394–8
Srikumar R, Li XZ, Poole K. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol 1997 Dec; 179(24): 7875–81
Li XZ, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol 1998 Jun; 180(11): 2987–91
Srikumar R, Kon T, Gotoh N, et al. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob Agents Chemother 1998 Jan; 42(1): 65–71
Li XZ, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can J Microbiol 1999 Jan; 45(1): 18–22
Masuda N, Sakagawa E, Ohya S, et al. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2000 Dec; 44(12): 3322–7
Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1990 Jan; 34(1): 52–7
Okamoto K, Gotoh N, Nishino T. Pseudomonas aeruginosa reveals high intrinsic resistance to penem antibiotics: penem resistance mechanisms and their interplay. Antimicrob Agents Chemother 2001 Jul; 45(7): 1964–71
Okamoto K, Gotoh N, Nishino T. Extrusion of penem antibiotics by multicomponent efflux systems MexAB-OprM, MexCD-OprJ, and MexXY-OprM of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2002 Aug; 46(8): 2696–9
Zhao Q, Li XZ, Srikumar R, et al. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother 1998 Jul; 42(7): 1682–8
Gotoh N, Tsujimoto H, Nomura A, et al. Functional replacement of OprJ by OprM in the MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa. FEMS Microbiol Lett 1998 Aug 1; 165(1): 21–7
Maseda H, Yoneyama H, Nakae T. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2000 Mar; 44(3): 658–64
Passador L, Cook JM, Gambello MJ, et al. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 1993 May 21; 260(5111): 1127–30
Hastings JW, Greenberg EP. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol 1999 May; 181(9): 2667–8
Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 1999 Feb; 181(4): 1203–10
Evans K, Passador L, Srikumar R, et al. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol 1998 Oct; 180(20): 5443–7
Hirakata Y, Srikumar R, Poole K, et al. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp Med 2002 Jul 1; 196(1): 109–18
Chuanchuen R, Beinlich K, Hoang TT, et al. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 2001 Feb; 45(2): 428–32
Morita Y, Komori Y, Mima T, et al. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol Lett 2001 Aug 7; 202(1): 139–43
Masuda N, Gotoh N, Ohya S, et al. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1996 Apr; 40(4): 909–13
Li XZ, Barre N, Poole K. Influence of the MexA-MexB-OprM multidrug efflux system on expression of the MexC-MexD-OprJ and MexE-MexF-OprN multidrug efflux systems in Pseudomonas aeruginosa. J Antimicrob Chemother 2000 Dec; 46(6): 885–93
Gotoh N, Tsujimoto H, Tsuda M, et al. Characterization of the MexC-MexD-OprJ multidrug efflux system in △MexA-MexB-OprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother 1998 Aug; 42(8): 1938–43
Masuda N, Sakagawa E, Ohya S, et al. Hypersusceptibility of the Pseudomonas aeruginosa nfxB mutant to β-lactams due to reduced expression of the AmpC β-lactamase. Antimicrob Agents Chemother 2001 Apr; 45(4): 1284–6
Ochs MM, McCusker MP, Bains M, et al. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother 1999 May; 43(5): 1085–90
Köhler T, van Delden C, Curty LK, et al. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol 2001 Sep; 183(18): 5213–22
Alonso A, Martinez JL. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 1997 May; 41(5): 1140–2
Zhang L, Li XZ, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother 2000 Feb; 44(2): 287–93
Alonso A, Martinez JL. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 2001 Jun; 45(6): 1879–81
Burns JL, Hedin LA, Lien DM. Chloramphenicol resistance in Pseudomonas cepacia because of decreased permeability. Antimicrob Agents Chemother 1989 Feb; 33(2): 136–41
Zhang L, Li XZ, Poole K. Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia. J Antimicrob Chemother 2001 Oct; 48(4): 549–52
Kim K, Lee S, Lee K, et al. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM 73. J Bacteriol 1998 Jul; 180(14): 3692–6
Sparling PF, Sarubbi Jr FA, Blackman E. Inheritance of low-level resistance to penicillin, tetracycline, and chloramphenicol in Neisseria gonorrhoeae. J Bacteriol 1975 Nov; 124(2): 740–9
Guymon LF, Sparling PF. Altered crystal violet permeability and lytic behavior in antibiotic-resistant and -sensitive mutants of Neisseria gonorrhoeae. J Bacteriol 1975 Nov; 124(2): 757–63
Lysko PG, Morse SA. Neisseria gonorrhoeae cell envelope: permeability to hydrophobic molecules. J Bacteriol 1981 Feb; 145(2): 946–52
Pan W, Spratt BG. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol 1994 Feb; 11(4): 769–75
Rouquette C, Harmon JB, Shafer WM. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol Microbiol 1999 Aug; 33(3): 651–8
Delahay RM, Robertson BD, Balthazar JT, et al. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 1997 Jul; 143 (Pt 7): 2127–33
Hagman KE, Lucas CE, Balthazar JT, et al. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 1997 Jul; 143 (Pt 7): 2117–25
Zarantonelli L, Borthagaray G, Lee EH, et al. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J Antimicrob Chemother 2001 May; 47(5): 651–4
McFarland L, Mietzner TA, Knapp JS, et al. Gonococcal sensitivity to fecal lipids can be mediated by an Mtr-independent mechanism. J Clin Microbiol 1983 Jul; 18(1): 121–7
Morse SA, Lysko PG, McFarland L, et al. Gonococcal strains from homosexual men have outer membranes with reduced permeability to hydrophobic molecules. Infect Immun 1982 Aug; 37(2): 432–8
Shafer WM, Veal WL, Lee EH, et al. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and N. imeningitidis. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 219–24
Garg P, Chakraborty S, Basu I, et al. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992–7 in Calcutta, India. Epidemiol Infect 2000 Jun; 124(3): 393–9
Huda MN, Morita Y, Kuroda T, et al. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a nonhalophilic bacterium. FEMS Microbiol Lett 2001 Sep 25; 203(2): 235–9
Colmer JA, Fralick JA, Hamood AN. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol Microbiol 1998 Jan; 27(1): 63–72
Baranwal S, Dey K, Ramamurthy T, et al. Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agents Chemother 2002 Aug; 46(8): 2676–8
Miyamae S, Ueda O, Yoshimura F, et al. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob Agents Chemother 2001 Dec; 45(12): 3341–6
Wigfield SM, Rigg GP, Kavari M, et al. Identification of an immunodominant drug efflux pump in Burkholderia cepacia. J Antimicrob Chemother 2002 Apr; 49(4): 619–24
Miyamae CC, Valvano MA. Cloning and characterization of the Burkholderia vietnamiensis norM gene encoding a multi-drug efflux protein. FEMS Microbiol Lett 2002 Oct 8; 215(2): 279–83
Nishino K, Yamaguchi A. Overexpression of the response regulator evgAof the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J Bacteriol 2001 Feb; 183(4): 1455–8
Naroditskaya V, Schlosser MJ, Fang NY, et al. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem Biophys Res Commun 1993 Oct 29; 196(2): 803–9
Phadtare S, Yamanaka K, Kato I, et al. Antibacterial activity of 4,5-dihydroxy-2-cyclopentan-1-one (DHCP) and cloning of a gene conferring DHCP resistance in Escherichia coli. J Mol Microbiol Biotechnol 2001 Jul; 3(3): 461–5
Yerushalmi H, Lebendiker M, Schuldiner S. EmrE, an Escherichia coli12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J Biol Chem 1995 Mar 24; 270(12): 6856–63
Turner RJ, Taylor DE, Weiner JH. Expression of Escherichia coliTehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother 1997 Feb; 41(2): 440–4
Li X-Z, Poole K, Nikaido H. Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosato aminoglycosides and dyes. Antimicrob Agents Chemother 2003; 47(1): 27–33
Hongo E, Morimyo M, Mita K, et al. The methyl viologen-resistance-encoding gene smvAof Salmonellatyphimurium. Gene 1994 Oct 11; 148(1): 173–4
Santiviago CA, Fuentes JA, Bueno SM, et al. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol Microbiol 2002 Nov; 46(3): 687–98
Baquero F. Gram-positive resistance: challenge for the development of new antibiotics. J Antimicrob Chemother 1997 May; 39 Suppl. A: 1–6
Hooper DC. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect Dis 2002 Sep; 2(9): 530–8
Ahmed M, Lyass L, Markham PN, et al. Two highly similar multidrug transporters of Bacillussubtiliswhose expression is differentially regulated. J Bacteriol 1995 Jul; 177(14): 3904–10
Baranova NN, Danchin A, Neyfakh AA. Mta, a global MerR-type regulator of the Bacillussubtilismultidrug-efflux transporters. Mol Microbiol 1999 Mar; 31(5): 1549–59
Woolridge DP, Vazquez-Laslop N, Markham PN, et al. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilismultidrug transporter Blt. J Biol Chem 1997 Apr 4; 272(14): 8864–6
Masaoka Y, Ueno Y, Morita Y, et al. A two-component multidrug efflux pump, EbrAB, in Bacillussubtilis. J Bacteriol 2000 Apr; 182(8): 2307–10
Davis DR, McAlpine JB, Pazoles CJ, et al. Enterococcus faecalismulti-drug resistance transporters: application for antibiotic discovery. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 179–84
Singh KV, Weinstock GM, Murray BE. An Enterococcusfaecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 2002 Jun; 46(6): 1845–50
Jonas BM, Murray BE, Weinstock GM. Characterization of emeA, a norAhomolog and multidrug resistance efflux pump, in Enterococcusfaecalis. Antimicrob Agents Chemother 2001 Dec; 45(12): 3574–9
Lynch C, Courvalin P, Nikaido H. Active efflux of antimicrobial agents in wild-type strains of enterococci. Antimicrob Agents Chemother 1997 Apr; 41(4): 869–71
Bolhuis H, van Veen HW, Molenaar D, et al. Multidrug resistance in Lactococcuslactis: evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J 1996 Aug 15; 15(16): 4239–45
Bolhuis H, Poelarends G, van Veen HW, et al. The lactococcal imrPgene encodes a proton motive force-dependent drug transporter. J Biol Chem 1995 Nov 3; 270(44): 26092–8
Perreten V, Schwarz FV, Teuber M, et al. Mdt (A), a new efflux protein conferring multiple antibiotic resistance in Lactococcus lactisand Escherichiacoli. Antimicrob Agents Chemother 2001 Apr; 45(4): 1109–14
Mata MT, Baquero F, Perez-Diaz JC. A multidrug efflux transporter in Listeriamonocytogenes. FEMS Microbiol Lett 2000 Jun 15; 187(2): 185–8
Ross JI, Eady EA, Cove JH, et al. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene 1995 Feb 3; 153(1): 93–8
Fournier B, Aras R, Hooper DC. Expression of the multidrug resistance transporter NorA from Staphylococcusaureusis modified by a two-component regulatory system. J Bacteriol 2000 Feb; 182(3): 664–71
Littlejohn TG, Pauken IT, Gillespie MT, et al. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcusaureus. FEMS Microbiol Lett 1992 Aug 15; 74(2–3): 259–65
Clancy J, Dib-Hajj F, Petitpas JW, et al. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob Agents Chemother 1997 Dec; 41(12): 2719–23
Gill MJ, Brenwald NP, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcuspneumoniae. Antimicrob Agents Chemother 1999 Jan; 43(1): 187–9
Tait-Kamradt A, Clancy J, Cronan M, et al. mefEis necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother 1997 Oct; 41(10): 2251–5
Clancy J, Petitpas J, Dib-Hajj F, et al. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcuspyogenes. Mol Microbiol 1996 Dec; 22(5): 867–79
Ainsa JA, Blokpoel MC, Otal I, et al. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacteriumfortuitumand Mycobacteriumtuberculosis. J Bacteriol 1998 Nov; 180(22): 5836–43
Takiff HE, Cimino M, Musso MC, et al. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacteriumsmegmatis. Proc Natl Acad Sci U S A 1996 Jan 9; 93(1): 362–6
Choudhuri BS, Sen S, Chakrabarti P. Isoniazid accumulation in Mycobacteriumsmegmatisis modulated by proton motive force-driven and ATP-dependent extrusion systems. Biochem Biophys Res Commun 1999 Mar 24; 256(3): 682–4
Choudhuri BS, Bhakta S, Barik R, et al. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrAand drrBof Mycobacterium tuberculosis. Biochem J 2002 Oct 1; 367 (Pt 1): 279–85
Doran JL, Pang Y, Mdluli KE, et al. Mycobacteriumtuberculosis efpAencodes an efflux protein of the QacA transporter family. Clin Diagn Lab Immunol 1997 Jan; 4(1): 23–32
Silva PE, Bigi F, de la Paz Santangelo M, et al. Characterization of P55, a multidrug efflux pump in Mycobacteriumbovisand Mycobacteriumtuberculosis. Antimicrob Agents Chemother 2001 Mar; 45(3): 800–4
De Rossi E, Branzoni M, Cantoni R, et al. mmr, a Mycobacterium tuberculosisgene conferring resistance to small cationic dyes and inhibitors. J Bacteriol 1998 Nov; 180(22): 6068–71
Carbon C. MRSA and MRSE: is there an answer? Clin Microbiol Infect 2000 Aug; 6 Suppl. 2: 17–22
Tennent JM, Lyon BR, Midgley M, et al. Physical and biochemical characterization of the qacAgene encoding antiseptic and disinfectant resistance in Staphylococcusaureus. J Gen Microbiol 1989 Jan; 135 (Pt 1): 1–10
Paulsen IT, Brown MH, Littlejohn TG, et al. Multidrug resistance proteins QacA and QacB from Staphylococcusaureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci U S A 1996 Apr 16; 93(8): 3630–5
Noguchi N, Hase M, Kitta M, et al. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcusaureus. FEMS Microbiol Lett 1999 Mar 15; 172(2): 247–53
Kaatz GW, Seo SM, Ruble CA. Efflux-mediated fluoroquinolone resistance in Staphylococcusaureus. Antimicrob Agents Chemother 1993 May; 37(5): 1086–94
Ng EY, Trucksis M, Hooper DC. Quinolone resistance mediated by norA: physiologic characterization and relationship toflqB, a quinolone resistance locus on the Staphylococcusaureus chromosome. Antimicrob Agents Chemother 1994 Jun; 38(6): 1345–55
Piddock LJ. Mechanisms of fluoroquinolone resistance: an update 1994–1998. Drugs 1999; 58 Suppl. 2: 11–8
Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother 2000; 44(9): 2233–41
Poole K. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob Agents Chemother 2000; 44(10): 2595–9
Yu JL, Grinius L, Hooper DC. NorA functions as a multidrug efflux protein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J Bacteriol 2002 Mar; 184(5): 1370–7
Kaatz GW, Seo SM. Inducible NorA-mediated multidrug resistance in Staphylococcusaureus. Antimicrob Agents Chemother 1995 Dec; 39(12): 2650–5
Fournier B, Truong-Bolduc QC, Zhang X, et al. A mutation in the 5' untranslated region increases stability of norAmRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J Bacteriol 2001 Apr; 183(7): 2367–71
Kaatz GW, Seo SM, Foster TJ. Introduction of a norApromoter region mutation into the chromosome of a fluoroquinolone-susceptible strain of Staphylococcusaureususing plasmid integration. Antimicrob Agents Chemother 1999 Sep; 43(9): 2222–4
Kaatz GW, Seo SM, O'Brien L, et al. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcusaureus. Antimicrob Agents Chemother 2000 May; 44(5): 1404–6
Munoz-Bellido JL, Alonzo Manzanares M, Martinez Andres JA, et al. Efflux pump-mediated quinolone resistance in Staphylococcusaureusstrains wild type for gyrA, gyrB, grlA, and norA. Antimicrob Agents Chemother 1999 Feb; 43(2): 354–6
Piddock LJ, Jin YF, Webber MA, et al. Novel ciprofloxacin-resistant, nalidixic acid-susceptible mutant of Staphylococcus aureus. Antimicrob Agents Chemother 2002 Jul; 46(7): 2276–8
Noguchi N, Tamura M, Narui K, et al. Frequency and genetic characterization of multidrug-resistant mutants of Staphylococcus aureusafter selection with individual antiseptics and fluoroquinolones. Biol Pharm Bull 2002 Sep; 25(9): 1129–32
Ross JI, Eady EA, Cove JH, et al. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol 1990 Jul; 4(7): 1207–14
Ross JI, Eady EA, Cove JH, et al. Minimal functional system required for expression of erythromycin resistance by msrAin StaphylococcusaureusRN 4220. Gene 1996 Dec 12; 183(1–2): 143–8
Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 2002 Feb 15; 34(4): 482–92
Schmitz FJ, Sadurski R, Kray A, et al. Prevalence of macrolide-resistance genes in Staphylococcusaureusand Enterococcus faeciumisolates from 24 European university hospitals. J Antimicrob Chemother 2000 Jun; 45(6): 891–4
Schmitz FJ, Perdikouli M, Beeck A, et al. Molecular surveillance of macrolide, tetracycline and quinolone resistance mechanisms in 1191 clinical European Streptococcuspneumoniae isolates. Int J Antimicrob Agents 2001 Nov; 18(5): 433–6
Broskey J, Coleman K, Gwynn MN, et al. Efflux and target mutations as quinolone resistance mechanisms in clinical isolates of Streptococcuspneumoniae. J Antimicrob Chemother 2000 Apr; 45 Suppl. 1: 95–9
Bast DJ, Low DE, Duncan CL, et al. Fluoroquinolone resistance in clinical isolates of Streptococcuspneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob Agents Chemother 2000 Nov; 44(11): 3049–54
Brenwald NP, Gill MJ, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcuspneumoniae. Antimicrob Agents Chemother 1998 Aug; 42(8): 2032–5
Baranova NN, Neyfakh AA. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother 1997 Jun; 41(6): 1396–8
Zeller V, Janoir C, Kitzis MD, et al. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother 1997 Sep; 41(9): 1973–8
Marshall NJ, Piddock LJ. Antibacterial efflux systems. Microbiologia 1997 Sep; 13(3): 285–300
Piddock LJ, Jin YF, Everett MJ. Non-gyrA-mediated ciprofloxacin resistance in laboratory mutants of Streptococcuspneumoniae. J Antimicrob Chemother 1997 May; 39(5): 609–15
Piddock LJ, Johnson MM. Accumulation of 10 fluoroquinolones by wild-type or efflux mutant Streptococcuspneumoniae. Antimicrob Agents Chemother 2002 Mar; 46(3): 813–20
Piddock LJ, Johnson MM, Simjee S, et al. Expression of efflux pump gene pmrAin fluoroquinolone-resistant and -susceptible clinical isolates of Streptococcuspneumoniae. Antimicrob Agents Chemother 2002 Mar; 46(3): 808–12
Pestova E, Millichap JJ, Siddiqui F, et al. Non-PmrA-mediated multidrug resistance in Streptococcuspneumoniae. J Antimicrob Chemother 2002 Mar; 49(3): 553–6
Kataja J, Seppala H, Skurnik M, et al. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob Agents Chemother 1998 Jun; 42(6): 1493–4
Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and p Syogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother 1996 Aug; 40(8): 1817–24
Widdowson CA, Klugman KP. Molecular mechanisms of resistance to commonly used non-betalactam drugs in Streptococcus pneumoniae. Semin Respir Infect 1999 Sep; 14(3): 255–68
French GL. Enterococci and vancomycin resistance. Clin Infect Dis 1998 Aug; 27 Suppl. 1: S75–83
Moellering Jr RC. Emergence of Enterococcus as a significant pathogen. Clin Infect Dis 1992 Jun; 14(6): 1173–6
Hallgren A, Abednazari H, Ekdahl C, et al. Antimicrobial susceptibility patterns of enterococci in intensive care units in Sweden evaluated by different MIC breakpoint systems. J Antimicrob Chemother 2001 Jul; 48(1): 53–62
Neyfakh AA. The ostensible paradox of multidrug recognition. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 151–4
Godsey MH, Baranova NN, Neyfakh AA, et al. Crystal structure of MtaN, a global multidrug transporter gene activator. J Biol Chem 2001 Dec 14; 276(50): 47178–84
Heldwein EE, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 2001 Jan 18; 409(6818): 378–82
Neyfakh AA. The multidrug efflux transporter of Bacillussubtilis is a structural and functional homolog of the Staphylococcus NorA protein. Antimicrob Agents Chemother 1992 Feb; 36(2): 484–5
Poelarends GJ, Mazurkiewicz P, Konings WN. Multidrug transporters and antibiotic resistance in Lactococcuslactis. Biochim Biophys Acta 2002 Sep 10; 1555(1–3): 1–7
van Veen HW, Putman M, Margolles A, et al. Molecular pharmacological characterization of two multidrug transporters in Lactococcuslactis. Pharmacol Ther 2000 Mar; 85(3): 245–9
van Veen HW, Venema K, Bolhuis H, et al. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR 1. Proc Natl Acad Sci U S A 1996 Oct 1; 93(20): 10668–72
van Veen HW, Margolles A, Muller M, et al. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J 2000 Jun 1; 19(11): 2503–14
Poelarends GJ, Konings WN. The transmembrane domains of the ABC multidrug transporter LmrA form a cytoplasmic exposed, aqueous chamber within the membrane. J Biol Chem 2002 Nov 8; 277(45): 42891–8
van Veen HW, Callaghan R, Soceneantu L, et al. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature 1998 Jan 15; 391(6664): 291–5
Hofmeyr JH, Rohwer JM, Snoep JL, et al. How to distinguish between the vacuum cleaner and flippase mechanisms of the LmrA multi-drug transporter in Lactococcuslactis. Mol Biol Rep 2002; 29(1–2): 107–12
Putman M, Van Veen HW, Degener JE, et al. Antibiotic resistance: era of the multidrug pump. Mol Microbiol 2000 May; 36(3): 772–3
Putman M, Koole LA, van Veen HW, et al. The secondary multidrug transporter LmrP contains multiple drug interaction sites. Biochemistry 1999 Oct 19; 38(42): 13900–5
Putman M, van Veen HW, Degener JE, et al. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology 2001 Oct; 147 (Pt 10): 2873–80
Dye C, Scheele S, Dolin P, et al. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO global surveillance and monitoring project. JAMA 1999; 282(7): 677–86
Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett 1994 Oct 15; 123(1–2): 11–8
Trias J, Benz R. Permeability of the cell wall of Mycobacterium smegmatis. Mol Microbiol 1994 Oct; 14(2): 283–90
Engelhardt H, Heinz C, Niederweis M. A tetrameric porin limits the cell wall permeability of Mycobacteriumsmegmatis. J Biol Chem 2002 Oct 4; 277(40): 37567–72
Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem 1995; 64: 29–63
Liu J, Takiff HE, Nikaido H. Active efflux of fluoroquinolones in Mycobacteriumsmegmatismediated by LfrA, a multidrug efflux pump. J Bacteriol 1996 Jul; 178(13): 3791–5
Sander P, De Rossi E, Boddinghaus B, et al. Contribution of the multidrug efflux pump LfrA to innate mycobacterial drug resistance. FEMS Microbiol Lett 2000 Dec 1; 193(1): 19–23
Wilson M, DeRisi J, Kristensen HH, et al. Exploring drug-induced alterations in gene expression in Mycobacteriumtuberculosis by microarray hybridization. Proc Natl Acad Sci U S A 1999 Oct 26; 96(22): 12833–8
Viveiros M, Portugal I, Bettencourt R, et al. Isoniazid-induced transient high-level resistance in Mycobacteriumtuberculosis. Antimicrob Agents Chemother 2002 Sep; 46(9): 2804–10
Zhang Y, Scorpio A, Nikaido H, et al. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacteriumtuberculosisto pyrazinamide. J Bacteriol 1999 Apr; 181(7): 2044–9
Schaller A, Guo M, Gisanrin O, et al. Escherichiacoligenes involved in resistance to pyrazinoic acid, the active component of the tuberculosis drug pyrazinamide. FEMS Microbiol Lett 2002 Jun 4; 211(2): 265–70
Piddock LJ, Williams KJ, Ricci V. Accumulation of rifampicin by Mycobacteriumaurum, Mycobacteriumsmegmatisand Mycobacterium tuberculosis. J Antimicrob Chemother 2000 Feb; 45(2): 159–65
Hui J, Gordon N, Kajioka R. Permeability barrier to rifampin in mycobacteria. Antimicrob Agents Chemother 1977 May; 11(5): 773–9
Li XZ, Wang YS, He ZN. Alteration of permeability of bacterial envelope barrier in rifamdin-resistant Mycobacteriumtuberculosis [in Chinese]. Hua Xi Yi Ke Da Xue Xue Bao 1988 Dec; 19(4): 388–91
Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacteriumtuberculosis. FEMS Microbial Rev 2000; 24(4): 449–67
Kaur P, Russell J. Biochemical coupling between the DrrA and DrrB proteins of the doxorubicin efflux pump of Streptomyces peucetius. J Biol Chem 1998 Jul 10; 273(28): 17933–9
Guilfoile PG, Hutchinson CR. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci U S A 1991 Oct 1; 88(19): 8553–7
Banerjee SK, Bhatt K, Misra P, et al. Involvement of a natural transport system in the process of efflux-mediated drug resistance in Mycobacteriumsmegmatis. Mol Gen Genet 2000 Jan; 262(6): 949–56
Bhatt K, Banerjee SK, Chakraborti PK. Evidence that phosphate specific transporter is amplified in a fluoroquinolone resistant Mycobacteriumsmegmatis. Eur J Biochem 2000 Jul; 267(13): 4028–32
Reizer J, Reizer A, Saier Jr MH. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci 1992 Oct; 1(10): 1326–32
Ninio S, Rotem D, Schuldiner S. Functional analysis of novel multidrug transporters from human pathogens. J Biol Chem 2001 Dec 21; 276(51): 48250–6
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacteriumtuberculosis: 1998 update. Tuber Lung Dis 1998; 79(1): 3–29
Zheleznova EE, Markham PN, Neyfakh AA, et al. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 1999 Feb 5; 96(3): 353–62
Vazquez-Laslop N, Markham PN, Neyfakh AA. Mechanism of ligand recognition by BmrR, the multidrug-responding transcriptional regulator: mutational analysis of the ligand-binding site. Biochemistry 1999 Dec 21; 38(51): 16925–31
Schumacher MA, Miller MC, Grkovic S, et al. Structural mechanisms of QacR induction and multidrug recognition. Science 2001 Dec 7; 294(5549): 2158–63
Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol 2002 Aug; 45(4): 885–93
Neyfakh AA. Mystery of multidrug transporters: the answer can be simple. Mol Microbiol 2002 Jun; 44(5): 1123–30
Vincent F, Spinelli S, Ramoni R, et al. Complexes of porcine odorant binding protein with odorant molecules belonging to different chemical classes. J Mol Biol 2000 Jun 30; 300(1): 127–39
Murakami S, Nakashima R, Yamashita E, et al. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002 Oct 10; 419(6907): 587–93
Fujihira E, Tamura N, Yamaguchi A. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichiacoli. J Biochem (Tokyo) 2002 Jan; 131(1): 145–51
Gotoh N, Kusumi T, Tsujimoto H, et al. Topological analysis of an RND family transporter, MexD of Pseudomonasaeruginosa. FEBS Lett 1999 Sep 10; 458(1): 32–6
Guan L, Ehrmann M, Yoneyama H, et al. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonasaeruginosa. J Biol Chem 1999 Apr 9; 274(15): 10517–22
Yoneyama H, Ocaktan A, Gotoh N, et al. Subunit swapping in the Mex-extrusion pumps in Pseudomonasaeruginosa. Biochem Biophys Res Commun 1998 Mar 27; 244(3): 898–902
Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric analysis of the multicomponent multidrug efflux transporters from gramnegative bacteria. J Bacteriol 2002 Dec; 184(23): 6499–507
Mao W, Warren MS, Black DS, et al. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonasaeruginosaare involved in substrate recognition. Mol Microbiol 2002 Nov; 46(3): 889–901
Yu EW, McDermott G, Zgurskaya HI, et al. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 2003; 300(5621): 976–80
Yu EW, Aires JR, Nikaido H. AcrB multidrug efflux pump of Escherichiacoli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J Bacteriol 2003; 185(19): 5657–64
Goldberg M, Pribyl T, Juhnke S, et al. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J Biol Chem 1999 Sep 10; 274(37): 26065–70
Guan L, Nakae T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonasaeruginosa. J Bacteriol 2001 Mar; 183(5): 1734–9
Aires JR, Pechere JC, Van Delden C, et al. Amino acid residues essential for function of the MexF efflux pump protein of Pseudomonasaeruginosa. Antimicrob Agents Chemother 2002 Jul; 46(7): 2169–73
Zgurskaya HI, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol 1999 Jan 8; 285(1): 409–20
Zgurskaya HI, Nikaido H. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J Bacteriol 2000 Aug; 182(15): 4264–7
Avila-Sakar AJ, Misaghi S, Wilson-Kubalek EM, et al. Lipid-layer crystallization and preliminary three-dimensional structural analysis of AcrA, the periplasmic component of a bacterial multidrug efflux pump. J Struct Biol 2001 Oct; 136(1): 81–8
Hayashi S, Wu HC. Lipoproteins in bacteria. J Bioenerg Biomembr 1990 Jun; 22(3): 451–71
Seiffer D, Klein JR, Plapp R. EnvC, a new lipoprotein of the cytoplasmic membrane of Escherichiacoli. FEMS Microbiol Lett 1993 Mar 1; 107(2–3): 175–8
Yoneyama H, Maseda H, Kamiguchi H, et al. Function of the membrane fusion protein, MexA, of the MexA,B-OprM efflux pump in Pseudomonasaeruginosawithout an anchoring membrane. J Biol Chem 2000 Feb 18; 275(7): 4628–34
Zgurskaya HI, Nikaido H. Multidrug resistance mechanisms: drug efflux across two membranes. Mol Microbiol 2000 Jul; 37(2): 219–25
Hwang J, Tai PC. Mutational analysis of CvaA in the highly conserved domain of the membrane fusion protein family. Curr Microbiol 1999 Oct; 39(4): 195–9
Hwang J, Zhong X, Tai PC. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J Bacteriol 1997 Oct; 179(20): 6264–70
Pimenta AL, Young J, Holland IB, et al. Antibody analysis of the localisation, expression and stability of HlyD, the MFP component of the E. colihaemolysin translocator. Mol Gen Genet 1999 Feb; 261(1): 122–32
Koronakis V, Sharff A, Koronakis E, et al. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 2000 Jun 22; 405(6789): 914–9
Andersen C, Hughes C, Koronakis V. Chunnel vision: export and efflux through bacterial channel-tunnels. EMBO Rep 2000 Oct; 1(4): 313–8
Wong KK, Brinkman FS, Benz RS, et al. Evaluation of a structural model of Pseudomonasaeruginosaouter membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. J Bacteriol 2001 Jan; 183(1): 367–74
Li XZ, Poole K. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonasaeruginosa. J Bacteriol 2001 Jan; 183(1): 12–27
Nakajima A, Sugimoto Y, Yoneyama H, et al. Localization of the outer membrane subunit OprM of resistance-nodulation-cell division family multicomponent efflux pump in Pseudomonas aeruginosa. J Biol Chem 2000 Sep 29; 275(39): 30064–8
Benz R, Maier E, Gentschev I. TolC of Escherichiacolifunctions as an outer membrane channel. Zentralbl Bakteriol 1993 Apr; 278(2–3): 187–96
Wong KK, Hancock RE. Insertion mutagenesis and membrane topology model of the Pseudomonasaeruginosaouter membrane protein OprM. J Bacteriol 2000 May; 182(9): 2402–10
Yoshihara E, Maseda H, Saito K. The outer membrane component of the multidrug efflux pump from Pseudomonasaeruginosa may be a gated channel. Eur J Biochem 2002 Oct; 269(19): 4738–45
Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr 1993 Dec; 25(6): 591–601
Zhao Q, Li XZ, Mistry A, et al. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonasaeruginosa. Antimicrob Agents Chemother 1998 Sep; 42(9): 2225–31
Godoy P, Ramos-Gonzalez MI, Ramos JL. Involvement of the TonB system in tolerance to solvents and drugs in Pseudomonas putidaDOT-T1E. J Bacteriol 2001 Sep; 183(18): 5285–92
Rouquette-Loughlin C, Stojiljkovic I, Hrobowski T, et al. Inducible, but not constitutive, resistance of gonococci to hydrophobic agents due to the MtrC-MtrD-MtrE efflux pump requires TonB-ExbB-ExbD proteins. Antimicrob Agents Chemother 2002 Feb; 46(2): 561–5
Maseda H, Kitao M, Eda S, et al. A novel assembly process of the multicomponent xenobiotic efflux pump in Pseudomonas aeruginosa. Mol Microbiol 2002 Nov; 46(3): 677–86
Helling RB, Janes BK, Kimball H, et al. Toxic waste disposal in Escherichiacoli. J Bacteriol 2002 Jul; 184(13): 3699–703
Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 2002 Dec; 66(4): 671–701
Ma D, Alberti M, Lynch C, et al. The local repressor AcrR plays a modulating role in the regulation of acrABgenes of Escherichia coliby global stress signals. Mol Microbiol 1996 Jan; 19(1): 101–12
Orth P, Schnappinger D, Hillen W, et al. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol 2000 Mar; 7(3): 215–9
Orth P, Cordes F, Schnappinger D, et al. Conformational changes of the Tet repressor induced by tetracycline trapping. J Mol Biol 1998 Jun 5; 279(2): 439–47
Bochner BR, Huang HC, Schieven GL, et al. Positive selection for loss of tetracycline resistance. J Bacteriol 1980 Aug; 143(2): 926–33
Masuda N, Sakagawa E, Ohya S, et al. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonasaeruginosa. Antimicrob Agents Chemother 2000 Sep; 44(9): 2242–6
Duque E, Segura A, Mosqueda G, et al. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonasputida. Mol Microbiol 2001 Feb; 39(4): 1100–6
Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichiacolimultidrug resistance pump EmrAB. J Bacteriol 1995 May; 177(9): 2328–34
Lomovskaya O, Kawai F, Matin A. Differential regulation of the mcband emroperons of Escherichiacoli: role of mcbin multidrug resistance. Antimicrob Agents Chemother 1996 Apr; 40(4): 1050–2
Relia M, Haas D. Resistance of PseudomonasaeruginosaPAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother 1982 Aug; 22(2): 242–9
Adewoye L, Sutherland A, Srikumar R, et al. The MexR repressor of the MexAB-OprM multidrug efflux operon in Pseudomonasaeruginosa:characterization of mutations compromising activity. J Bacteriol 2002 Aug; 184(15): 4308–12
Saito K, Yoneyama H, Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexRgene of the Pseudomonasaeruginosa chromosome. FEMS Microbiol Lett 1999 Oct 1; 179(1): 67–72
Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonasaeruginosa. Microb Drug Resist 1998 Winter; 4(4): 257–61
Evans K, Adewoye L, Poole K. MexR repressor of the MexAB-OprM multidrug efflux operon of Pseudomonasaeruginosa: identification of MexR binding sites in the MexA-MexR intergenic region. J Bacteriol 2001 Feb; 183(3): 807–12
Lim D, Poole K, Strynadka NC. Crystal structure of the MexR repressor of the mexRAB-oprMmultidrug efflux operon of Pseudomonasaeruginosa. J Biol Chem 2002 Aug 9; 277(32): 29253–9
Alekshun MN, Levy SB, Mealy TR, et al. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat Struct Biol 2001 Aug; 8(8): 710–4
Cao L, Srikumar R, Poole K. Identification and characterization of nalCmultidrug-resistant isolates of Pseudomonasaeruginosa [abstract no. C1-430]. Abstracts of the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, American Society for Microbiology, Washington, DC; 2002 Sep 27–30, San Diego (CA), 29
Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem 1992 Aug 5; 267(22): 15869–74
Shiba T, Ishiguro K, Takemoto N, et al. Purification and characterization of the PseudomonasaeruginosaNfxB protein, the negative regulator of the nfxBgene. J Bacteriol 1995 Oct; 177(20): 5872–7
Köhler T, Epp SF, Curty LK, et al. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonasaeruginosa. J Bacteriol 1999 Oct; 181(20): 6300–5
Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol 1993; 47: 597–626
Grkovic S, Brown MH, Roberts NJ, et al. QacR is a repressor protein that regulates expression of the Staphylococcusaureus multidrug efflux pump QacA. J Biol Chem 1998 Jul 17; 273(29): 18665–73
Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem 2000; 69: 183–215
Kieboom J, Dennis JJ, Zylstra GJ, et al. Active efflux of organic solvents by PseudomonasputidaS12 is induced by solvents. J Bacteriol 1998 Dec; 180(24): 6769–72
Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the marregulon. Antimicrob Agents Chemother 1997 Oct; 41(10): 2067–75
Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to maroperator sequences. Proc Natl Acad Sci U S A 1995 Jun 6; 92(12): 5456–60
Seoane AS, Levy SB. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol 1995 Jun; 177(12): 3414–9
Gallegos MT, Michan C, Ramos JL. The XylS/AraC family of regulators. Nucleic Acids Res 1993 Feb 25; 21(4): 807–10
Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichiacoliby constitutive expression of MarA. J Bacteriol 2000 Jun; 182(12): 3467–74
Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichiacoli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 1996 Jan; 178(1): 306–8
Li H, Park JT. The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichiacoli. J Bacteriol 1999 Aug; 181(16): 4842–7
Ma D, Cook DN, Alberti M, et al. Genes acrAand acrBencode a stress-induced efflux system of Escherichiacoli. Mol Microbiol 1995 Apr; 16(1): 45–55
McMurry LM, Oethinger M, Levy SB. Overexpression of marA, soxS, or acrABproduces resistance to triclosan in laboratory and clinical strains of Escherichiacoli. FEMS Microbiol Lett 1998 Sep 15; 166(2): 305–9
White DG, Goldman JD, Demple B, et al. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robAin Escherichiacoli. J Bacteriol 1997 Oct; 179(19): 6122–6
Hidalgo E, Ding H, Demple B. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem Sci 1997 Jun; 22(6): 207–10
Kwon HJ, Bennik MH, Demple B, etal. Crystal structure of the Escherichia coliRob transcription factor in complex with DNA. Nat Struct Biol 2000 May; 7(5): 424–30
Nakajima H, Kobayashi K, Kobayashi M, etal. Overexpression of the robAgene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol 1995 Jun; 61(6): 2302–7
Ariza RR, Li Z, Ringstad N, et al. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coliRob protein. J Bacteriol 1995 Apr; 177(7): 1655–61
Jair KW, Yu X, Skarstad K, et al. Transcriptional activation of promoters of the Superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol 1996 May; 178(9): 2507–13
Rosenberg EY, Bertenthal D, Nilles ML, et al. Bile salts and fatty acids induce the expression of Escherichia coliAcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 2003; 48(6): 1609–19
Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coliK-12. J Bacteriol 1998 Feb; 180(4): 938–44
Cohen SP, McMurry LM, Levy SB. marAlocus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol 1988 Dec; 170(12): 5416–22
Rosner JL, Chai TJ, Foulds J. Regulation of ompFporin expression by salicylate in Escherichia coli. J Bacteriol 1991 Sep; 173(18): 5631–8
Nikaido H, Rosenberg EY, Foulds J. Porin channels in Escherichia coli: studies with β-lactams in intact cells. J Bacteriol 1983 Jan; 153(1): 232–40
Nikaido H, Rosenberg EY. Porin channels in Escherichiacoli: studies with liposomes reconstituted from purified proteins. J Bacteriol 1983 Jan; 153(1): 241–52
Rahmati S, Yang S, Davidson AL, et al. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol Microbiol 2002 Feb; 43(3): 677–85
George AM, Hall RM, Stokes HW. Multidrug resistance in Klebsiellapneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 1995 Aug; 141 (Pt 8): 1909–20
Vaara M. Antibiotic-supersusceptible mutants of Escherichia coliand Salmonellatyphimurium. Antimicrob Agents Chemother 1993 Nov; 37(11): 2255–60
Mazzariol A, Cornaglia G, Nikaido H. Contributions of the AmpC β-lactamase and the AcrAB multidrug efflux system in intrinsic resistance of Escherichia coliK-12 to β-lactams. Antimicrob Agents Chemother 2000 May; 44(5): 1387–90
Cho D, Blais J, Tangen K, et al. Prevalence of efflux pumpsamong clinical isolates of fluoroquinolone-resistant Pseudomonas aeruginosa[abstract no. 1267]. Abstracts of the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Washington, DC; 1999 Sep 26–29; San Francisco (CA), 327
George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichiacoli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol 1983 Aug; 155(2): 531–40
George AM, Levy SB. Gene in the major cotransduction gap of the Escherichia coliK-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol 1983 Aug; 155(2): 541–8
Russell AD. Do biocides select for antibiotic resistance? J Pharm Pharmacol 2000 Feb; 52(2): 227–33
Cohen SP, Levy SB, Foulds J, et al. Salicylate induction of antibiotic resistance in Escherichia coli:activation of the mar operon and a mar-independent pathway. J Bacteriol 1993 Dec; 175(24): 7856–62
Sumita Y, Fukasawa M. Transient carbapenem resistance induced by salicylate in Pseudomonasaeruginosaassociated with suppression of outer membrane protein D2 synthesis. Antimicrob Agents Chemother 1993 Dec; 37(12): 2743–6
Burns JL, Clark DK. Salicylate-inducible antibiotic resistance in Pseudomonascepaciaassociated with absence of a poreforming outer membrane protein. Antimicrob Agents Chemother 1992 Oct; 36(10): 2280–5
Domenico P, Hopkins T, Cunha BA. The effect of sodium salicylate on antibiotic susceptibility and synergy in Klebsiella pneumoniae. J Antimicrob Chemother 1990 Sep; 26(3): 343–51
Schaller A, Sun Z, Yang Y, et al. Salicylate reduces susceptibility of Mycobacteriumtuberculosisto multiple antituberculosis drugs. Antimicrob Agents Chemother 2002 Aug; 46(8): 2636–9
Price CT, O'Brien FG, Shelton BP, et al. Effects of salicylate and related compounds on fusidic acid MICs in Staphylococcus aureus. J Antimicrob Chemother 1999 Jul; 44(1): 57–64
Gustafson JE, Candelaria PV, Fisher SA, et al. Growth in the presence of salicylate increases fluoroquinolone resistance in Staphylococcusaureus. Antimicrob Agents Chemother 1999 Apr; 43(4): 990–2
Price CT, Kaatz GW, Gustafson JE. The multidrug efflux pump NorA is not required for salicylate-induced reduction in drug accumulation by Staphylococcusaureus. Int J Antimicrob Agents 2002 Sep; 20(3): 206–13
Williams RJ, Livermore DM, Lindridge MA, et al. Mechanisms of β-lactam resistance in British isolates of Pseudomonas aeruginosa. J Med Microbiol 1984 Jun; 17(3): 283–93
Bert F, Lambert-Zechovsky N. Comparative distribution of resistance patterns and serotypes in Pseudomonasaeruginosa isolates from intensive care units and other wards. J Antimicrob Chemother 1996 Apr; 37(4): 809–13
Jakics EB, Iyobe S, Hirai K, et al. Occurrence of the nfxBtype mutation in clinical isolates of Pseudomonasaeruginosa. Antimicrob Agents Chemother 1992 Nov; 36(11): 2562–5
Fukuda H, Hosaka M, Iyobe S, et al. nfxC-type quinolone resistance in a clinical isolate of Pseudomonasaeruginosa. Antimicrob Agents Chemother 1995 Mar; 39(3): 790–2
Beinlich KL, Chuanchuen R, Schweizer HP. Contribution of multidrug efflux pumps to multiple antibiotic resistance in veterinary clinical isolates of Pseudomonasaeruginosa. FEMS Microbiol Lett 2001 May 1; 198(2): 129–34
Hoiby N. New antimicrobials in the management of cystic fibrosis. J Antimicrob Chemother 2002 Feb; 49(2): 235–8
Charvalos E, Tselentis Y, Hamzehpour MM, et al. Evidence for an efflux pump in multidrug-resistant Campylobacterjejuni. Antimicrob Agents Chemother 1995 Sep; 39(9): 2019–22
Mazzariol A, Tokue Y, Kanegawa TM, et al. High-level fluoroquinolone-resistant clinical isolates of Escherichia colioverproduce multidrug efflux protein AcrA. Antimicrob Agents Chemother 2000 Dec; 44(12): 3441–3
Wang H, Dzink-Fox JL, Chen M, et al. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrRmutations. Antimicrob Agents Chemother 2001 May; 45(5): 1515–21
George AM. Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol Lett 1996 May 15; 139(1): 1–10
Linde HJ, Notka F, Irtenkauf C, et al. Increase in MICs of ciprofloxacin invivoin two closely related clinical isolates of Enterobactercloacae. J Antimicrob Chemother 2002 Apr; 49(4): 625–30
Deguchi T, Kawamura T, Yasuda M, et al. Invivoselection of Klebsiellapneumoniaestrains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob Agents Chemother 1997 Jul; 41(7): 1609–11
del Mar Tavio M, Vila J, Ruiz J, et al. Decreased permeability and enhanced proton-dependent active efflux in the development of resistance to quinolones in Morganellamorganii. Int J Antimicrob Agents 2000 Mar; 14(2): 157–60
Ishii H, Sato K, Hoshino K, et al. Active efflux of ofloxacin by a highly quinolone-resistant strain of Proteusvulgaris. J Antimicrob Chemother 1991 Dec; 28(6): 827–36
Ishida H, Fuziwara H, Kaibori Y, et al. Cloning of multidrug resistance gene pqrAfrom Proteusvulgaris. Antimicrob Agents Chemother 1995 Feb; 39(2): 453–7
Ghosh AS, Ahamed J, Chauhan KK, et al. Involvement of an efflux system in high-level fluoroquinolone resistance of Shigelladysenteriae. Biochem Biophys Res Commun 1998 Jan 6; 242(1): 54–6
Perez-Trallero E, Fernandez-Mazarrasa C, Garcia-Rey C, et al. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenesisolates and their ecological relationships: results of a 1-year (1998–1999) multi-center surveillance study in Spain. Antimicrob Agents Chemother 2001 Dec; 45(12): 3334–40
Levy SB. Active efflux, a common mechanism for biocide and antibiotic resistance. J Appl Microbiol 2002; 92 Suppl.: 65S–71S
Lambert RJ, Joynson J, Forbes B. The relationships and susceptibilities of some industrial, laboratory and clinical isolates of Pseudomonasaeruginosato some antibiotics and biocides. J Appl Microbiol 2001 Dec; 91(6): 972–84
Stickler DJ. Susceptibility of antibiotic-resistant Gram-negative bacteria to biocides: a perspective from the study of catheter biofilms. J Appl Microbiol 2002; 92 Suppl.: 163S–70S
Higgins CS, Murtough SM, Williamson E, et al. Resistance to antibiotics and biocides among non-fermenting Gram-negative bacteria. Clin Microbiol Infect 2001 Jun; 7(6): 308–15
Nakahara H, Asakawa M, Yonekura I, et al. Benzethonium chloride resistance in Pseudomonasaeruginosaisolated from clinical lesions. Zentralbl Bakteriol Mikrobiol Hyg [A] 1984 Aug; 257(3): 409–13
Nakahara H, Kozukue H. Isolation of chlorhexidine-resistant Pseudomonasaeruginosafrom clinical lesions. J Clin Microbiol 1982 Jan; 15(1): 166–8
Block C, Furman M. Association between intensity of chlorhexidine use and micro-organisms of reduced susceptibility in a hospital environment. J Hosp Infect 2002 Jul; 51(3): 201–6
Fraise AP. Biocide abuse and antimicrobial resistance: a cause for concern? J Antimicrob Chemother 2002 Jan; 49(1): 11–2
Skurray RA, Rouch DA, Lyon BR, et al. Multiresistant Staphylococcusaureus: genetics and evolution of epidemic Australian strains. J Antimicrob Chemother 1988 Apr; 21 Suppl. C: 19–39
Lyon BR, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 1987 Mar; 51(1): 88–134
Join-Lambert OF, Michea-Hamzehpour M, Kohler T, et al. Differential selection of multidrug efflux mutants by trovafloxacin and ciprofloxacin in an experimental model of Pseudomonasaeruginosaacute pneumonia in rats. Antimicrob Agents Chemother 2001 Feb; 45(2): 571–6
Köhler T, Michea-Hamzehpour M, Plesiat P, et al. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1997 Nov; 41(11): 2540–3
Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis 2000 Aug; 31 Suppl. 2: S24–8
Zhao X, Drlica K. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis 2001 Sep 15; 33 Suppl. 3: S147-56
Mamber SW, Kolek B, Brookshire KW, et al. Activity of quinolones in the Ames SalmonellaTA102 mutagenicity test and other bacterial genotoxicity assays. Antimicrob Agents Chemother 1993 Feb; 37(2): 213–7
Ysern P, Clerch B, Castano M, et al. Induction of SOSgenes in Escherichia coliand mutagenesis in Salmonellatyphimurium by fluoroquinolones. Mutagenesis 1990 Jan; 5(1): 63–6
Le Thomas I, Couetdic G, Clermont O, et al. Invivoselection of a target/efflux double mutant of Pseudomonasaeruginosaby ciprofloxacin therapy. J Antimicrob Chemother 2001 Oct; 48(4): 553–5
Zhanel GG, Karlowsky JA, Saunders MH, et al. Development of multiple-antibiotic-resistant (Mar) mutants of Pseudomonas aeruginosaafter serial exposure to fluoroquinolones. Antimicrob Agents Chemother 1995 Feb; 39(2): 489–95
Lomovskaya O, Lee A, Hoshino K, et al. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonasaeruginosa. Antimicrob Agents Chemother 1999 Jun; 43(6): 1340–6
Lee A, Mao W, Warren MS, et al. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 2000 Jun; 182(11): 3142–50
Plesiat P, Nikaido H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol Microbiol 1992 May; 6(10): 1323–33
Vaara M. The outer membrane as the penetration barrier against mupirocin in gram-negative enteric bacteria. J Antimicrob Chemother 1992 Feb; 29(2): 221–2
Yethon JA, Gunn JS, Ernst RK, et al. Salmonellaenterica serovar typhimurium waaPmutants show increased susceptibility to polymyxin and loss of virulence invivo. Infect Immun 2000 Aug; 68(8): 4485–91
Yethon JA, Heinrichs DE, Monteiro MA, et al. Involvement of waaY, waaQ, and waaPin the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem 1998 Oct 9; 273(41): 26310–6
Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev 1992 Sep; 56(3): 395–411
Fralick JA, Burns-Keliher LL. Additive effect of tolCand rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coliK-12. J Bacteriol 1994 Oct; 176(20): 6404–6
Lucas CE, Hagman KE, Levin JC, et al. Importance of lipooligosaccharide structure in determining gonococcal resistance to hydrophobic antimicrobial agents resulting from the mtr efflux system. Mol Microbiol 1995 Jun; 16(5): 1001–9
Li XZ, Nikaido H, Williams KE. Silver-resistant mutants of Escherichia colidisplay active efflux of Ag+ and are deficient in porins. J Bacteriol 1997 Oct; 179(19): 6127–32
Nikaido H, Normark S. Sensitivity of Escherichia colito various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol Microbiol 1987 Jul; 1(1): 29–36
Li XZ, Zhang L, Poole K. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonasaeruginosa. J Antimicrob Chemother 2000 Apr; 45(4): 433–6
Plesiat P, Aires JR, Godard C, et al. Use of steroids to monitor alterations in the outer membrane of Pseudomonasaeruginosa. J Bacteriol 1997 Nov; 179(22): 7004–10
Germ M, Yoshihara E, Yoneyama H, et al. Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonasaeruginosa. Biochem Biophys Res Commun 1999 Aug 2; 261(2): 452–5
Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonasaeruginosa:our worst nightmare? Clin Infect Dis 2002 Mar 1; 34(5): 634–40
Lakaye B, Dubus A, Lepage S, et al. When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Mol Microbiol 1999 Jan; 31(1): 89–101
Nakae T, Nakajima A, Ono T, et al. Resistance to β-lactam antibiotics in Pseudomonasaeruginosadue to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob Agents Chemother 1999 May; 43(5): 1301–3
Masuda N, Gotoh N, Ishii C, et al. Interplay between chromosomal β-lactamase and the MexAB-OprM efflux system in intrinsic resistance to β-lactams in Pseudomonasaeruginosa. Antimicrob Agents Chemother 1999 Feb; 43(2): 400–2
Srikumar R, Tsang E, Poole K. Contribution of the MexAB-OprM multidrug efflux system to the β-lactam resistance of penicillin-binding protein and β-lactamase-derepressed mutants of Pseudomonasaeruginosa. J Antimicrob Chemother 1999 Oct; 44(4): 537–40
Li XZ, Zhamg L, McKay GA, et al. Role of the acetyltransferase AAC(6')-Iz modifying enzyme in aminoglycoside resistance in Stenotrophomonasmaltophilia. J Antimicrob Chemother 2003; 51(4): 803–11
Kaatz GW, Seo SM. Mechanisms of fluoroquinolone resistance in genetically related strains of Staphylococcusaureus. Antimicrob Agents Chemother 1997 Dec; 41(12): 2733–7
Nakajima A, Sugimoto Y, Yoneyama H, et al. High-level fluoroquinolone resistance in Pseudomonasaeruginosadue to interplay of the MexAB-OprM efflux pump and the DNA gyrase mutation. Microbiol Immunol 2002; 46(6): 391–5
Veal WL, Nicholas RA, Shafer WM. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrRmutation is required for chromosomally mediated penicillin resistance in Neisseriagonorrkoeae. J Bacteriol 2002 Oct; 184(20): 5619–24
Maiti SN, Phillips OA, Micetich RG, et al. Beta-lactamase inhibitors: agents to overcome bacterial resistance. Curr Med Chem 1998 Dec; 5(6): 441–56
Chopra I. New developments in tetracycline antibiotics: glycylcyclines and tetracycline efflux pump inhibitors. Drug Resist Updat 2002 Aug 6; 5 (3–4): 119
Nelson ML, Levy SB. Reversal of tetracycline resistance mediated by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob Agents Chemother 1999 Jul; 43(7): 1719–24
Nelson ML, Park BH, Levy SB. Molecular requirements for the inhibition of the tetracycline antiport protein and the effect of potent inhibitors on the growth of tetracycline-resistant bacteria. J Med Chem 1994 Apr 29; 37(9): 1355–61
Rothstein DM, McGlynn M, Bernan V, et al. Detection of tetracyclines and efflux pump inhibitors. Antimicrob Agents Chemother 1993 Aug; 37(8): 1624–9
Nelson ML. Modulation of antibiotic efflux in bacteria. Curr Med Chem-Anti-Infective Agents 2002; 1(1): 35–54
Hirata T, Wakatabe R, Nielsen J, et al. A novel compound, 1,1-dimethyl-5 (1-hydroxypropyl)-4,6,7-trimethylindan, is an effective inhibitor of the tet(K@#@)gene-encoded metal-tetracycline/ H+ antiporter of Staphylococcusaureus. FEBS Lett 1997 Jul 28; 412(2): 337–40
Hirata T, Wakatabe R, Nielsen J, et al. Screening of an inhibitor of the tetracycline efflux pump in a tetracycline-resistant clinical-isolate of Staphylococcusaureus743. Biol Pharm Bull 1998 Jul; 21(7): 678–81
Barrett JF. MC-207110 Daiichi Seiyaku/Microcide Pharmaceuticals. Curr Opin Investig Drugs 2001 Feb; 2(2): 212–5
Ryan BM, Dougherty TJ, Beaulieu D, et al. Efflux in bacteria: what do we really know about it? Expert Opin Investig Drugs 2001 Aug; 10(8): 1409–22
Lewis K. In search of natural substrates and inhibitors of MDR pumps. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 247–54
Wigler PW, Patterson FK. Inhibition of the multidrug resistance efflux pump. Biochim Biophys Acta 1993 Oct 29; 1154(2): 173–81
Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodiumfalciparumby verapamil. Science 1987 Feb 20; 235(4791): 899–901
Cohn RC, Rudzienski L, Putnam RW. Verapamil-tobramycin synergy in Pseudomonascepaciabut not Pseudomonas aeruginosainvitro. Chemotherapy 1995 Sep–Oct; 41(5): 330–3
Brenwald NP, Gill MJ, Wise R. The effect of reserpine, an inhibitor of multi-drug efflux pumps, on the in-vitrosusceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniaeto norfloxacin. J Antimicrob Chemother 1997 Sep; 40(3): 458–60
Gibbons S, Udo EE. The effect of reserpine, a modulator of multidrug efflux pumps, on the invitroactivity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus(MRSA) possessing the tet(K@#@)determinant. Phytother Res 2000 Mar; 14(2): 139–40
Beyer R, Pestova E, Millichap JJ, et al. A convenient assay for estimating the possible involvement of efflux of fluoroquino-lones by Streptococcus pneumoniaeand Staphylococcusaureus: evidence for diminished moxifloxacin, sparfloxacin, and trovafloxacin efflux. Antimicrob Agents Chemother 2000 Mar; 44(3): 798–801
Markham PN, Westhaus E, Klyachko K, et al. Multiple novel inhibitors of the NorA multidrug transporter of Staphylococcus aureus. Antimicrob Agents Chemother 1999 Oct; 43(10): 2404–8
Neyfakh AA, Borsch CM, Kaatz GW. Fluoroquinolone resistance protein NorA of Staphylococcusaureusis a multidrug efflux transporter. Antimicrob Agents Chemother 1993 Jan; 37(1): 128–9
Stermitz FR, Lorenz P, Tawara JN, et al. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci U S A 2000 Feb 15; 97(4): 1433–7
Renau TE, Leger R, Flamme EM, et al. Inhibitors of efflux pumps in Pseudomonasaeruginosapotentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem 1999 Dec 2; 42(24): 4928–31
Renau TE, Leger R, Flamme EM, et al. Addressing the stability of C-capped dipeptide efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonasaeruginosa. Bioorg Med Chem Lett 2001 Mar 12; 11(5): 663–7
Lomovskaya O, Warren MS, Lee A, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonasaeruginosa:novel agents for combination therapy. Antimicrob Agents Chemother 2001 Jan; 45(1): 105–16
Griffith D, Lomovskaya O, Lee V, et al. Potentiation of levofloxacin by a broad-spectrum efflux inhibitor (EPI) in mouse models of infection caused by Pseudomonasaeruginosa [abstract no. 1268]. Abstracts of the 39th Interscience conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Washington, DC; 1999 Sep 26–29; San Francisco (CA), 327
Renau TE, Leger R, Yen R, et al. Peptidomimetics of efflux pump inhibitors potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett 2002 Mar 11; 12(5): 763–6
Lee MD, Galazzo JL, Staley AL, et al. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 2001 Jan–Feb; 56(1–2): 81–5
Fukuda H, Hori S, Hiramatsu K. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norAtransformant of Staphylococcus aureus. Antimicrob Agents Chemother 1998 Aug; 42(8): 1917–22
Gootz TD, Zaniewski RP, Haskell SL, et al. Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrAand grlAmutants of Staphylococcusaureus. Antimicrob Agents Chemother 1999 Aug; 43(8): 1845–55
Ince D, Hooper DC. Mechanisms and frequency of resistance to premafloxacin in Staphylococcusaureus: novel mutations suggest novel drug-target interactions. Antimicrob Agents Chemother 2000 Dec; 44(12): 3344–50
Zhong P, Shortridge VD. The role of efflux in macrolide resistance. Drug Resist Updat 2000 Dec; 3(6): 325–9
Chu DT. Recent progress in novel macrolides, quinolones, and 2-pyridones to overcome bacterial resistance. Med Res Rev 1999 Nov; 19(6): 497–520
Brennan L, Duignan J, Petitpas J, et al. CP-544372: MIC90 studies and killing kinetics against key respiratory tract pathogens [abstract no. F-124]. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Washington, DC; 1998 Sep 24–27; San Diego (CA), 264
Someya Y, Yamaguchi A, Sawai T. A novel glycylcycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, is neither transported nor recognized by the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Antimicrob Agents Chemother 1995 Jan; 39(1): 247–9
Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A 1973 Aug; 70(8): 2276–80
Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994 Apr 15; 264(5157): 375–82
Davies J. Another look at antibiotic resistance: 1991 Fred Griffith Review Lecture. J Gen Microbiol 1992 Aug; 138 (Pt 8): 1553–9
Marshall CG, Lessard IA, Park I, et al. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother 1998 Sep; 42(9): 2215–20
Krulwich TA, Jin J, Guffanti AA, et al. Functions of tetracycline efflux proteins that do not involve tetracycline. J Mol Microbiol Biotechnol 2001 Apr; 3(2): 237–46
Wang W, Guffanti AA, Wei Y, et al. Two types of Bacillus subtilisTetA(L @#@)deletion strains reveal the physiological importance of TetA(L @#@)in K+ acquisition as well as in Na+, alkali, and tetracycline resistance. J Bacteriol 2000 Apr; 182(8): 2088–95
Acknowledgements
Research in the authors’ laboratory was supported by a grant from the US Public Health Service (AI-09644). A portion of the review is based on the PhD thesis of X.-Z. Li, who thanks Keith Poole for his encouragement during the graduate study in his laboratory. Neither author has any disclosable interest relevant to the content of the review.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, XZ., Nikaido, H. Efflux-Mediated Drug Resistance in Bacteria. Drugs 64, 159–204 (2004). https://doi.org/10.2165/00003495-200464020-00004
Published:
Issue Date:
DOI: https://doi.org/10.2165/00003495-200464020-00004