Abstract
The insulinlike growth factor-I (IGF-I) is an important factor which regulates a variety of cellular responses in multiple biological systems. The IGF1 gene comprises a highly conserved sequence and contains six exons, which give rise to heterogeneous mRNA transcripts by a combination of multiple transcription initiation sites and alternative splicing. These multiple transcripts code for different precursor IGF-I polypeptides, namely the IGF-IEa, IGF-IEb and IGF-IEc isoforms in humans, which also undergo posttranslational modifications, such as proteolytic processing and glycosylation. IGF-I actions are mediated through its binding to several cell-membrane receptors and the IGF-I domain responsible for the receptor binding is the bioactive mature IGF-I peptide, which is derived after the posttranslational cleavage of the pro-IGF-I isoforms and the removal of their carboxy-terminal E-peptides (that is, the Ea, Eb and Ec). Interestingly, differential biological activities have been reported for the different IGF-I isoforms, or for their E-peptides, implying that IGF-I peptides other than the IGF-I ligand also possess bioactivity and, thus, both common and unique or complementary pathways exist for the IGF-I isoforms to promote biological effects. The multiple peptides derived from IGF-I and the differential expression of its various transcripts in different conditions and pathologies appear to be compatible with the distinct cellular responses observed to the different IGF-I peptides and with the concept of a complex and possibly isoform-specific IGF-I bioactivity. This concept is discussed in the present review, in the context of the broad range of modifications that this growth factor undergoes which might regulate its mechanism(s) of action.
Similar content being viewed by others
Introduction
The insulinlike growth factor-I (IGF-I), also called somatomedin C, is a cellular and secreted growth factor which is critical for normal body growth, development and maintenance, and has important roles in multiple biological systems (1–3). A variety of cellular responses are induced by IGF-I, including cell proliferation, differentiation, migration and survival (4–8). These cellular responses have implicated IGF-I in several conditions such as the pathophysiology of several cancers (9–11), or the mitogenic and myogenic processes during muscle development, regeneration or hypertrophy, since, unlike other growth factors, IGF-I acts as both a mitogen and a differentiation factor (12,13).
IGF-I is produced by many tissues, indicating that a significant component of IGF-I action is due to its autocrine and paracrine mode of function, although it also acts as a classical circulating hormone. In the endocrine mode of action, IGF-I acts as a mediator (somatomedin C) of the growth-promoting effects of pituitary growth hormone (GH, somatotropin), which induces the synthesis and release of IGF-I by the liver (2,14,15). Circulating IGF-I is mainly derived from the liver, but also from skeletal muscle (3,16–18), and is mostly bound to high affinity IGF-binding proteins, which protect it from proteolytic degradation and modulate its bioavailability to the IGF-I receptors (2,19).
Different IGF-I mRNA transcripts are produced as a result of the alternative splicing of the IGF1 gene, encoding for several IGF-I precursor proteins. These IGF-I protein isoforms differ by the structure of their extension peptides, or E-peptides, on the carboxy-terminal end and by the length of their amino-terminal signal peptides. However, they share the same mature peptide, which is the common part of all the IGF-I precursors (20–23). IGF-I mediates its actions through the binding and activation of several receptors and the IGF-I domain which is responsible for the receptor binding is the biologically active mature peptide. It is derived after the posttranslational cleavage of the pro-IGF-I (iso)forms and the removal of the E-peptides (23–26). Interestingly, it has been proposed that the E-peptides also possess bioactivity that is distinct from that of mature IGF-I (20,27).
Thus, during the last decade, many in vitro and in vivo studies have investigated the aspect of the differential IGF-I isoforms or their E-peptides actions in various conditions and pathologies (28–38). This concept was further supported by recent findings which revealed differential, E-peptide- or IGF-I isoform-specific signaling (31,33,34,38–40).
In the present review, focus has been on the propounded concept of the differential roles and bioactivity of the IGF-I isoforms or peptides, in the context of the complexity that characterizes the alternative splicing, posttranscriptional regulation and posttranslational modifications of this growth factor and which might modulate its mechanism(s) of action.
Human IGF1 Gene Structure and Alternative Splicing
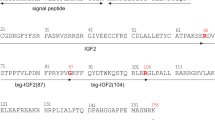
The IGF1 gene spans a region of over 80 kb of genomic DNA located on the long arm of chromosome 12 in humans, it is a highly conserved sequence in mammals and primates (23), and contains six exons, which give rise to heterogeneous mRNA transcripts by a combination of multiple transcription initiation sites (that is, alternative leader sequences), alternative splicing and different polyadenylation signals (41,42). These multiple IGF-I transcripts code different precursor polypeptides, which also undergo posttranslational modifications (26,43,44), (Figures 1A–C).
Gene structure, alternative splicing, amino acid sequence and posttranslational modifications of human IGF-I. Posttranslational cleavage of pro-IGF-I polypeptides can occur at a unique pentabasic motif and at sites containing the general PC consensus sequence (panels 1A–C; potential processing sites are indicated by arrows; see text for details). Only the Ea-peptide of human IGF-I contains an N-linked glycosylation site. Mature IGF-I peptide residues that interact with IGFBPs are shown in orange color and underlined whereas residues that interact with IGF-IR are in purple color and marked with an asterisk (*) (panel 1A).
More specifically, the different leader sequences result in two different classes of IGF-I mRNA variants: class 1 transcripts have their initiation sites on exon 1 (promoter 1), whereas class 2 transcripts use exon 2 as leader exon (promoter 2), and class 1 (exon 1 to exon 3) or class 2 (exon 2 to the exon 3) mRNA transcripts are produced by differential splicing of exons 1 and 2 to the common exon 3. Alternative splicing of exon 5 also results in different mRNA variants containing exon 5, generally defined as class B (IGF-IEb), or containing exon 6 (and excluding exon 5) defined as class A (IGF-IEa) (26,45,46), (see Figures 1A, B). A third variant, the IGF-IEc, which corresponds to IGF-IEb in rodents, also is generated by alternative splicing in the human IGF1 gene and contains both exon 5 and 6 (22), (see Figure 1C). Similar to the human IGF1 gene, multiple forms of pro-IGF-I mRNA have been described in other species in which IGF-I genomic sequences have been determined, such as the designated pro-IGF-I Ea-1, Ea-2, Ea-3 and Ea-4 in teleosts (23,47–50).
All possible combinations between promoter usage and terminal exon (5 or 6) can occur in different IGF-I transcripts (45,51,52). It has been proposed that the use of promoter 1 could be associated with the synthesis of paracrine IGF-I and may influence interactions with insulinlike growth factor binding proteins (IGF-BPs), or promote the formation of the truncated IGF-I peptide (46), (see below: IGF-I Processing, Secretion and Glycosylation). Transcripts initiating at promoter 1 are widely expressed in many tissues, whereas transcripts initiating at promoter 2 are expressed mainly in the liver (circulating forms) and kidney (53) and are thought to be more GH-dependent (23,54–58), or equally GH-responsive (59,60). However, the two promoters are probably not mutually exclusive, and GH can also stimulate the expression of tissue-specific (local) transcripts, although the existent evidence is still equivocal (48,61–65).
IGF-IEa transcript derives from the splicing pattern exon 1 or 2–3–4–6 of the IGF1 gene, which represents the main pro-IGF-I mRNA produced in liver (systemic IGF-IEa) but also in other tissues with similar exon sequence (22,66), (see Figure 1A). IGF-IEb transcript is a splice variant of exon 1 or 2–3–4–5. Its expression was firstly detected in the human liver (67), while it was also found to be expressed in lung carcinoma cells (20), in skeletal muscle (31,62,68) and more recently in various tissues and cells such as prostate, endometrium and lens epithelial cells (34,35,65,69), (see Figure 1B). Whether there is equivalent to human IGF-IEb splice pattern in nonhuman primates is not known (23,43,70). IGF-IEc mRNA transcript is an exon 1 or 2–3–4–5–6 splice variant (see Figure 1C), which was initially identified also in human liver, where, however, it is expressed approximately at 10% relative to the main IGF-IEa transcript (22). Structurally, its cDNA differs from the IGF-IEa variant by the presence of the first 49 base pairs from exon 5 (52 bp in rodents), It results from a splice acceptor site in the intron preceding exon 6 and, due to a reading frame shift, it gives rise to a different carboxy-terminal peptide sequence and a premature stop codon in exon 6. This transcript was named mechano-growth factor (MGF) since it was found to be upregulated in response to muscle stretch and/or damage (71); for review see (72). However, its expression also has been identified in various tissues such as endometrium (35), normal and cancerous prostatic cells (34), as well as in osteoblast-like osteosarcoma cells (73).
The biological significance of IGF-I splice variants is currently unknown and the physiological and molecular mechanisms that regulate their expression are unclear; however, the presence of distinct transcripts is indicative of diverse responses of cells to different stimuli (74) and they probably reflect the complexity of IGF-I actions mediated via its various isoforms (75,76).
Recent studies in humans have shown that the IGF-I splice variants are differentially transcribed in response to varying conditions and pathologies, such as exercise-induced muscle damage (31,68), endometriosis (35), and prostate (34), or cervical cancer (69), as well as in some human cell lines after hormonal treatment (65,73), while their differential transcription is possibly a function of age (64,72). The differential expression of the IGF-I splice variants observed in various pathologies is of particular interest, as it could indicate distinct regulatory mechanisms and biological roles of the different IGF-I isoforms; however, their particular functions remain as yet unclear.
The different IGF-I mRNA transcripts encode the corresponding precursor proteins IGF-IEa, IGF-IEb and IGF-IEc (23,44). The 5′ end, by alternative splicing of exons 1, 2 and 3, encodes for the signal peptide of the IGF-I prohormone. Four different transcription start sites are present in exon 1 and their positions relative to the translational initiation codons (that is, Met-48 located in exon 1, and Met-25 and Met-22 located in exon 3) can give rise to three distinct IGF-I signal peptides from class 1 mRNAs (exon 1 to exon 3), (23,43). However, translation of mRNAs initiated at the four transcription start sites mentioned above is expected to produce signal sequences of 48 (transcription start sites 1, 2 and 3) and 25 (transcription start site 4) amino acids, since between two translation start sites contained in an mRNA, (for example, Met-25 and Met-22, located downstream of the transcription start site 4), the preference is given to the upstream site (42,44,55,66), (see Figures 1A–C). It was suggested that when translation initiates at Met-25, the nucleotide sequence corresponding to the first 21 amino acids (encoded by exon 1) of the sequence of the 48 amino acids (aa) signal peptide may have a specific function as part of the promoter 1, or may play a role in posttranscriptional regulation of IGF-I mRNA by GH (23,55,58).
From class 2 (exon 2 to exon 3) transcripts, three transcription start sites and their upstream position relative to the translational initiation codon Met-32 (located in exon 2) give rise to an IGF-I precursor polypeptide with 32 aa long signal sequence (23,42,44), (see Figures 1A–C).
The mature IGF-I peptide is coded by exons 3 (25 aa) and 4 (45 aa). The first 16 amino acids of the amino-terminal portion of the IGF-I E-peptide are coded by exon 4. Exons 5 and 6 encode, by alternative splicing, distinct portions of the E-peptide with alternative carboxyterminal sequences that contain also distinct termination codons (22,44). Three different E-peptides have been identified in humans, encoded by three mRNA variants produced by alternative splicing of the 3′ end of the pre-IGF-I mRNA. Exon 4 to exon 6 mRNA splicing encodes the Ea-peptide, which contains 35 aa. The first 16 aa, which are common in all the E-peptides, are encoded by the exon 4 and the remaining 19 are encoded by exon 6 (44,66). Splice variant of exon 4 to exon 5 yields the Eb-peptide which, apart from the 16 common aa encoded by the exon 4, contains 61 additional aa encoded by exon 5, resulting in the 77 aa long Eb-peptide (44,67). The third mRNA splice variant, which contains exon 4, only 49 bp from exon 5, and then exon 6, produces the Ecpeptide with a predicted length of 40 aa, that is, 16 aa from the exon 4, 16 aa from the exon 5 and 8 aa from the exon 6 (22,44), (see Figures 1A–C). It is noted, that the last 8 aa of Ec-peptide are encoded by exon 6, however they differ from the corresponding Ea-peptide sequence, because of a frameshift at the splice point. Ec-peptide is thought to occur by use of a cryptic IGF633 donor splice site, which is located 49 bp downstream from the 5′ end of the exon 5. When this cryptic IGF633 donor splice site is not used, the alternative splicing of exon 4–5 occurs, that is, the Eb peptide (22,43). The predicted molecular mass and the residues of the three different isoforms of the human IGF-I precursor polypeptide as well as of their various forms and partial peptides (based on the amino acid sequence derived from the various IGF-I mRNA transcripts) are summarized in Table 1.
In general, the complexity introduced by the transcriptional and splicing variants, posttranscriptional regulation and posttranslational modifications of the IGF1 gene (77), giving rise to various IGF-I isoforms, probably indicate their different biological roles under various conditions or pathologies and following different stimuli (78). The development of epitope-specific antibodies for distinguishing the different IGF-I E-peptides (20,38,79–82) can contribute to a more definitive analysis of IGF-I isoforms expression in various tissues and physiological or pathophysiological conditions.
IGF-I Processing, Secretion and Glycosylation
Post translational processing of IGF-I precursor protein may be a regulatory mechanism of the IGF-I activity, as indicated by the unique processing features of IGF-I precursor polypeptides that have been described (25,78). Posttranslational endoproteolysis of those polypeptides produces the signal, the mature and the E-peptides (E domains). It is thought that the signal peptide at the start of a precursor is removed after facilitating the passage of the polypeptide into the endoplasmic reticulum and the secretory pathway, with possibly no further biological significance (23,83). The sequences of the signal peptides and the E-peptides are less strongly conserved compared with mature IGF-I peptide, though to a variable extent (23).
The mature peptide comprises four domains, that is, the B amino-terminal domain, C and A domain and D carboxy-terminal domain, of IGF-I polypeptides (25,84). In addition, two other protein products have been identified in the human brain; the tripeptide glycyl-prolyl-glutamate (GPE) corresponding to the NH2-terminal of the B domain of mature IGF-I and a truncated IGF-I form (−3N:IGF-I) that lacks the first three amino acids of the amino terminal end of mature peptide, probably due to alternate signal peptides or the combined action of some peptidases (78,85,86). Removal of the NH2-terminal tripeptide could be a mechanism for increasing the biological potency and availability of IGF-I, since the truncated −3N:IGF-I has less affinity for IGF-binding proteins than mature IGF-I, thus, increasing its bioactivity (78), (see Figure 1A) (see below: IGF-I Receptors and Binding Proteins).
The mature IGF-I is a 70 aa long single-chain peptide and a highly conserved sequence among primate species (23,24). Cleavage of pro-IGF-I removes the carboxyl-terminal E domain and can occur at the highly conserved, unique pentabasic motif K65-X-X-K68-X-X-R71-X-X-R74-X-X-R77. More specifically, the Arg71-Ser72 bond is cleaved followed by the removal of the Arg residue by the action a carboxypeptidase (25), (see Figures 1A–C). In general, proproteins can be processed at this specific motif, usually residing at the end of their pro regions, by proprotein convertases (PCs) such as furin (25,87,88). Furin belongs to the subtilisin-related PCs (SPCs), a major family of endoproteolytic processing enzymes of the secretory pathway in mammals (89,90). Seven mammalian PCs have been identified, namely PC1, PC2, furin, PC4, PC5, paired basic amino acid cleaving enzyme 4 (PACE4) and PC7, and a method of prediction of the general PC-specific or furin-specific cleavage sites has been proposed (88). PCs process precursors at sites usually containing the specific consensus sequence [R/K]-Xn-[R/K], where X indicates any amino acid residue, and n, the number of spacer amino acid residues, which is 0, 2, 4 or 6 (91), (see Figures 1A–C). However, furin appears to have a more stringent specificity and preferentially recognizes sites that contain the sequence motif R-X-[R/K]-R (87), while R-X-X-R is its minimal cleavage sequence (88). Thus, apart from the cleavage site Arg71-Ser72 for mature IGF-I, another furin-mediated processing at Arg77 has been observed in overexpression studies, which produces an extended, 76 aa long mature IGF-I (25,78), (see Figures 1A–C).
PC-mediated processing of pro-IGF-I to mature peptide has been shown to occur intracellularly (92), as expected for intracellular convertases such as furin, which are located in the secretory pathway (92,93). Nevertheless, evidence has been provided that the E domains are not cleaved intracellularly (94), and the secretion of unprocessed pro-IGF-IEa isoform, both glycosylated and nonglycosylated, has been reported (25,30,95–97), while there are potential proprotein convertases that could process pro-IGF-I extracellularly (97,98).
Conversion of pro-IGF-I to mature peptide cleaves off the E domains of the IGF-I precursors and one peptide is produced from the human Ea domain after posttranslational processing of pro-IGF-IEa isoform (23,80), (see Figure 1A). However, the Eb domain contains potential processing sites (see Figure 1B), and at least two putative peptide products of the human Eb peptide have been identified: the EB1 peptide (residues 103–124 aa) with a C-terminal amide and the EB2 peptide amide (residues 129–142) (20). The Ec domain also contains potential processing sites, from which one corresponds to the EB1 peptide cleavage site and cleaves off the last eight residues of the Ec domain encoded by the exon 6 (26,82), (see Figures 1B, C). However, so far only the proform of IGF-IEc isoform and not the Ec peptide or other cleavage products have been detected (31),33–35,73,82).
The Ea-peptide of human IGF-I contains an N-linked glycosylation site at Asn92 based on the consensus sequence Asn-X-Ser/Thr, where X represents any encoded amino acid except proline (99,100), (see Figure 1A). However, it has been found that the human IGF-IEa precursor with a signal sequence of 48 aa (Met-48) is not glycosylated, implying that the greater the number of the amino acids contained in the signal peptide the lesser the extent of a glycosylation process (101). The human Eb- and Ec-peptides lack this cotranslational modification due to the reading frame shift resulted from the inclusion of the exon 5 in these two IGF-I isoforms (see Figures 1B, C). N-linked glycosylation involves the transfer of a lipid-linked tetradecasaccharide (GlcNAc2-Man9-Glc3) to an asparagine side chain (100,102), (see Figure 1A). It occurs in endoplasmic reticulum and the subsequent diversification of the conjugates occurs both in the endoplasmic reticulum and Golgi apparatus (100,102). Recently, it has been found that the glycosylation status of pro-IGF-IEa does not affect its processing (97), while the secretion of a peptide similar or identical to Ea from a human B-lymphocyte cell line, in a partially glycosylated form has been reported (80). Although the deglycosylation process is expected to occur intracellularly, whether the deglycosylation of pro-IGF-IEa or the Ea-peptide also could be clipped extracellularly is not known.
Considering the unique role of glycosylation in the protein biosynthesis process (100), it is possible that the Ea-peptide glycosylation might play a role in interactions with chaperones in the endoplasmic reticulum (78), or in regulation of the bioavailability of the different species of this IGF-I isoform (that is, pro-IGF-I Ea, mature IGF-I or Ea-peptide) (78,97,103). Thus, the existence of an N-linked glycosylation site in the Ea-peptide, which is absent in the Ec- and Eb-peptide, might reflect a differential and specific biological action of the IGF-IEa isoform mediated by this posttranslational modification of its Ea-peptide (25,43,97). Recently, it has been shown that the species of IGF-I produced by the IGF-IEa isoform has a differential ability to activate IGF-IR. Glycosylated pro-IGF-IEa is less efficient at receptor activation than pro-IGF-I and mature IGF-I (97), resembling the decreased receptor-binding affinity of pegylated IGF-I forms (104,105) and implying that glycosylated pro-IGF-IEa may serve as a reservoir for IGF-I that can be stored until needed (97). The strong conservation observed in the sequence of the Ea domain also suggests a specific biological function for the Ea-peptide (23).
Nevertheless, whether the E-peptides are more stable and/or bioactive within their pro-IGF-I forms, or they are processed to act directly on their targets remains to be elucidated (see further discussion in next sections). Moreover, it would be essential to verify whether the IGF-I isoforms could be released in the circulation as different proforms or E-peptides (3,79), or the final peptide that enters the circulation after extracellular endoproteolysis of the IGF-I prohormone is only the mature peptide (103,106).
IGF-I Receptors and Binding Proteins
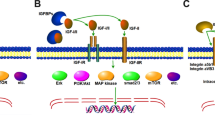
IGF-I actions are mediated through its binding to several receptors, such as IGF-IR (or type I IGF receptor) and IGF-IIR (or type II IGF receptor), insulin receptor (IR), and some atypical receptors such as the hybrid IR/IGF-IR (107–109).
More specifically, the mature IGF-I peptide, which is responsible for binding to the receptors, binds IGF-IR with the highest affinity, IGF-IIR with low affinity and is also able to interact with IR. The IGF-IR exhibits a high degree of homology to IR (110) and, given the significant structural similarity between IGF-I and insulin, these ligands can cross-activate both receptors, while the IGF-IR signaling pathways share multiple intracellular mediators with the insulin signaling cascade (19,52). The IGF-IR/IR hybrid receptor is thought to function predominantly as an IGF-I receptor, since its binding affinity for insulin is lower than that for IGF-I, however the functional importance of IGF-IR/IR hybrid receptor remains poorly understood (74,109,111).
It is widely recognized that most of the observed IGF-I biological effects on cell growth, differentiation, invasion and survival depend on the binding and activation of IGF-IR, which is a ligand-activated receptor tyrosine kinase (4,112). Specifically, functional epitope mapping of IGF-I has revealed that the IGF-IR interacts with the residues 21, 23, 24, 44 as well as the tyrosines 31 and 60, which are located in the C and A domains (113), (see Figure 1A). IGF-IR is a transmembrane protein consisting of two extracellular α-subunits, which contain the cysteine-rich ligand binding site, and two transmembrane β-subunits that have a cluster of three tyrosine residues, which undergo phosphorylation and activation upon IGF-I binding (2,110,114). A structural rearrangement in the transmembrane β subunits of the receptor is caused by binding of IGF-I to IGF-IR, resulting in transautophosphorylation of the cytoplasmic tyrosine kinase domain of the receptor, as one kinase domain phosphorylates the other, and thus destabilizing the autoinhibitory conformation within the kinase domain (115,116). This conformational change permits unrestricted access to the binding sites for protein substrates (117), thus recruiting specific cytoplasmic molecules, such as insulin receptor substrate (IRS) proteins, and activating specific intracellular pathways including Ras/mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinases 1 and 2 (ERK1/2) and phosphatidylinositol 3-kinase (PI3K)/Akt (118).
Biological actions of IGF-I are modulated by a family of at least six IGFBPs (52,75,119–21), which interact mainly with the residues 1–3 and 49–51 of mature IGF-I (113), (see Figure 1A). In general, IGFBPs transport IGF-I and increase its half-life in the circulation; most of the circulating IGF-I is protected from proteolytic degradation by forming a ternary complex with IGFBP-3 and the glycoprotein acid-labile subunit (ALS) (74,122). IGFBPs also would be expected to modulate and control, both in the circulation and in the extracellular environment, the extent of IGF-dependent cellular effects via regulation of free IGF-I concentration and its local bioavailability in the tissue, since IGF-BPs provide tissue specificity for the local action of IGF-I (74,119,123–126). In addition, IGFBPs compete with IGF-IR and normally have higher binding affinity to IGF-I than IGF-IR does. Therefore, binding of IGFBPs to IGF-I prevents the ligand from interacting with the receptor and, thus, suppresses IGF-I actions (74,124,125).
However, some IGFBPs can exhibit IGF-I potentiating effects and their IGF-I-inhibitory or stimulatory activities are dictated by factors such as the tissue-specific distribution of particular IGFBPs and the ratio between free (active) IGF-I and IGFBP-IGF-I bound (4,11). Moreover, it has been shown that certain IGFBPs have IGF-independent activities, implying that they can modulate cell survival and apoptosis, or inhibit tumor growth in the absence of the ligand (11,127). In addition, proteolytic fragments of IGFBP-3 were reported to possess mitogenic activity in the peritoneal fluid of women with endometriosis (128).
There is also a group of cysteine-rich proteins, known as IGFBP-related proteins (IGFBP-rPs), that share important structural similarities with the IGFBPs but they have low binding affinity to IGFs. It has been proposed that these proteins and the IGFBPs constitute an IGFBP superfamily (129,130), however the functions of the IGFBP-rPs regarding the IGFs actions are as yet unclear (131).
IGF-I Peptides Actions and Signaling
Although, by the general consensus, IGF-I is thought to exert its biological actions predominantly through mature peptide, differential biological activities have been reported for the different IGF-I isoforms (propeptides), or for their E-peptides, exogenously administrated or overexpressed in various in vivo (28,29,36,132–134) and in vitro models (20),27,31–33,135,136), implying that there are peptides other than the IGF-I ligand that also possess bioactivity and, thus, both common and unique or complementary pathways exist for the IGF-I isoforms to promote biological effects (31,36).
A differential expression profile of the IGF-I isoforms have been shown in various conditions or pathologies in humans, such as skeletal muscle damage (31,68), endometriosis (35) or prostate (34), cervical (69) and colorectal cancer (137). The diversity and the patterns of differential expression have been proposed to reflect potential biological activities associated with the E domain peptides (27,52). Divergent actions and signaling of the different pro-IGF-I forms or mature IGF-I lacking any E-peptide have been reported after viral-mediated expression of the IGF-I isoforms (IGF-IA and IGF-IB) in mouse skeletal muscle (36,132). Specifically, it has been shown that overexpression of mature IGF-I in skeletal muscle did not promote muscle hypertrophy in young mice, suggesting that the pro-IGF-I forms are required for this effect and that E-peptides, either as a part of pro-IGF-I or independent of mature IGF-I, may be necessary for IGF-I-mediated muscle hypertrophy (36). Moreover, by using the MKR transgenic mouse model where functional IGF-IRs are lacking in muscle fibers (as a dominant negative IGF-IR is expressed specifically in skeletal muscle), it was shown that, regardless of which isoform is overexpressed, IGF-I receptors on muscle fibers are required for IGF-I-mediated hypertrophy (36). Interestingly, however, after viral-mediated delivery of murine IGF-I isoforms into skeletal muscle, even though both isoforms caused increased phosphorylation of the IGF-IR, increased expression of IGF-IB (murine IGF-IEb) drove both main pathways downstream of IGF-IR, that is, the PI3K/Akt pathway and the MAPK pathway, whereas IGF-IA (murine IGF-IEa) overexpression resulted in increased Akt phosphorylation only (132). These findings imply distinguishing IGF-I isoform-specific actions, regardless of any potential receptor(s) activated (see also discussion in following sections). In addition, constitutively overexpression of the IGF-IEa isoform specifically in cardiac muscle was shown to protect the heart from oxidative stress via Sirtuin 1 (SirT1)/c-Jun N-terminal kinase 1 (JNK1) activity while, conversely, mature IGF-I triggered oxidative stress in the heart and did not affect SirT1 activity (138,139). Moreover, it has been shown that although both mature IGF-I and IGF-IEa propeptide triggered the phosphorylation of IGF-IR, its downstream canonical PI3K/Akt/mTOR signaling pathway was not induced in the transgenic mice overexpressing IGF-IEa (138,140). Instead, this specific IGF-I propeptide activated alternate signaling intermediates 3-phosphoinositide-dependent protein kinase-1 (PDK1) and serum/glucocorticoid regulated kinase 1 (SGK1), as well as SirT1. Thus, it was suggested that this downstream of IGF-I receptor(s) signaling activated by IGF-IEa employs novel pathways and that the divergent signaling mechanisms between the two IGF-I forms (that is, mature IGF-I and IGF-IEa propeptide) may account for their opposing effects on the heart (136,138,140).
Considering particularly the bioactivity of the E-peptides, in vitro studies have suggested that the E-peptides of the human IGF-I precursors may act as independent growth factors, since their synthetic analogs, generated from unique regions within the E domains, were demonstrated to possess mitogenic (20,27,34,35,73,141), angiogenic (142) and migratory activity (28,141,143), and regulate cell differentiation (27,28) in various human cells or cell lines. Antitumor activity of human Eb-peptide also has been reported in some cancer cells (144).
Similar to the results derived from human cell lines, studies using animal cell lines models have shown that exogenous administration or overexpression of synthetic peptides, generated from different regions within the E domain of human (Ec) (28,31,33,145,146), trout (Ea) (144,147) or rodent (Ea and Eb) sequences (38) in human, rodent or porcine cells in culture showed unique though inconsistent effects in promoting cell proliferation and migration, and in delaying or inhibiting cell differentiation. In particular, actions associated with peptides derived from mammalian Ea domain have only recently been established, with rodent Ea-peptide reported to possess bioactivity (38). Furthermore, it was shown that murine Ea- and Eb-peptides increase cell entry of IGF-I from the media, providing evidence that, in addition to having independent activity, they may modulate IGF-I (30).
The differential biological effects of the synthetic Ec-peptide compared with mature IGF-I peptide, such as cell proliferation versus differentiation, and the lack of suppression of the synthetic E-peptide bioactivity after blocking (mature) IGF-I signaling with IGF-IR neutralizing antibodies, makes it tempting to postulate that the Ec-peptide acts via a different receptor (28,135,145). However, concerns have been raised about the effectiveness of the IGF-IR neutralizing antibodies to block IGF-I signaling, since they could internalize and, in this way, even activate the receptor, or change its localization, thus facilitating an E-peptide action (38). Nevertheless, the mature IGF-I bioactivity appeared to be suppressed in those cells where IGF-IR neutralizing antibodies were used.
Interestingly, an autonomous, IGF-IR-independent bioactivity has been reported for specific regions of the human Eb domain of IGF-I; the synthetic analog of the EB1 peptide (see above: IGF-I Processing, Secretion and Glycosylation) was found not only to possess mitogenic activity in human bronchial epithelial cells but also to bind to specific high-affinity receptors on those cells. Furthermore, neither ligand binding was inhibited by recombinant IGF-I or recombinant insulin, nor did a monoclonal antibody antagonist to the IGF-IR suppress the proliferative response induced by the synthetic EB1 peptide. It was suggested that IB1 is a growth factor that mediates its effect through a specific receptor (20). Similarly, in accordance with the well established notion that the molecular action of a bioactive peptide initiates through its specific binding to a cell surface receptor, it was demonstrated that the human Eb-peptide binds to common cell surface molecules on human neuroblastoma cells (27). Again, the saturation of mature IGF-I or insulin did not displace the binding of the human Eb-peptide, suggesting the existence of distinct putative receptor components on those cells (148).
Further evidence for distinct, IGF-IR-independent bioactivity of the human Ec domain was provided from its divergent signaling compared with mature IGF-I. Our group (31,33,34,65) and others (39) have shown that a synthetic analog of the human Ec peptide possesses distinct signaling compared with IGF-IR ligand. Specifically, to distinguish any unique biological effect of the Ec domain from a potential bioactivity possessed by the common part (that is, the first 16 residues) of the IGF-I E domains, we utilized a synthetic Ec-peptide corresponding to the region beyond that common sequence, that is, a peptide similar to the C-terminal 24-residues, of which 16 are encoded by exon 5 and the last 8 by exon 6. It was documented that this sequence of the human Ec domain possesses distinct signaling since, in contrast to mature IGF-I, it activates only ERK1/2 and not Akt (31,33,34,65). Moreover, the selective activation of only one of the two main signaling pathways downstream of IGF-I/IGF-IR and the IGF-IR- and IR- independent bioactivity of this synthetic part of the Ec domain was further documented by using the siRNA knock-out model in various human cell lines (34,35,73).
More recently, synthetic E-peptides corresponding to the rodent Ea and Eb domain sequences were utilized in a kinase receptor activation (KIRA) assay to test IGF-I-dependent and -independent activation of IGF-IR by these E-peptides, showing that they do not directly induce IGF-IR phosphorylation in mouse fibroblasts (P6 cells) (38). Interestingly, however, the presence of either of those E-peptides increased IGF-IR activation by IGF-I in the murine C2C12 cell line, suggesting that they may modulate IGF-I activity (38). Moreover, from the findings of that model, it was concluded that E-peptides signaling, as well as mitogenic and motogenic effects are dependent upon a functional IGF-IR, and their activity plausibly reflects actions of pro-IGF-I (38). Further evidence has been provided recently, suggesting that IGF-I splice variants may exert their actions through mature IGF-I and not the E-peptides (149), or by supporting the bioactivity of pro-IGF-I forms (97). Furthermore, there is controversial evidence regarding the role of proforms (103,136,149) or E-peptides (38) of the murine IGF-I isoforms, particularly in cell differentiation. Whether it reflects a differential mode of action of the free E-peptides compared with the pro-IGF-I isoforms, or potential differences between their exogenous administration and overexpression (or natural endogenous production), possibly should be addressed.
Collectively, the in vitro models utilizing the sequences of the rodent IGF-I E domains have provided specific information regarding the bioactivity and the mode of action of those E-peptides in murine cell lines. The findings of these studies suggest that the E-peptides have little or no independent activity and, instead, they modulate mature IGF-I (ligand) activity and signaling (see review in [150]). Nevertheless, since these domains are very variable and much less conserved, it has been indicated that the species specificity must clearly be taken into account when assessing the activity of the human IGF-I Eb and Ec domains, from which peptides with important biological activities have been reported (23). Thus, it remains to be further elucidated whether the autonomous, IGF-IR-independent and IR-independent bioactivity, and the specific high-affinity receptor binding observed for human Eb- and Ec-peptides in various human cell lines (20,34,35,73,148) reflect an alternative, species-specific ligand/receptor mechanism of action for these human E domains. Species-specific models, in terms of utilizing peptide sequences that correspond to the IGF-I E domains of the species or the cells used in the model, are of particular importance to investigate physiological mechanisms of actions, such as a ligand/receptor mechanism.
Moreover, it is still an elusive fact where the E-peptides signaling diverges from that of mature IGF-I (given the distinct activation of ERK1/2 but not Akt pathway by the E-peptide). It has been proposed that the E-peptides may increase the phosphorylation of ERK1/2 by increasing the IGF-IR internalization, which would act in favor of the MAPK arm of the IGF-IR downstream signaling (38). However, since the possibility that the E-peptides also activate ERK1/2 through an IGF-IR independent mechanism cannot be excluded, it remains to be further elucidated whether they affect the ERK1/2 pathway only at the level of the IGF-IR per se (that is, possibly by increasing its internalization, or by affecting its conformational change after its binding to IGF-I and, thus, activating exclusively a specific pathway) or if the E-peptide-induced activation of ERK1/2 occurs at a level downstream of the IGF-IR, or even upstream, in that the E-peptide utilizes another “receptor” or cellular uptake mechanism, given the activation of ERK1/2 after exogenous administration of the E-peptide. The intracellular signaling pathways initiated by IGF-IR ligation may interact with signaling via G protein-coupled receptors or other mediators, modulating some responses (151). However, the existence of such a putative, canonical or noncanonical receptor or internalization mechanism for the E-peptide remains to be determined and characterized. Alternatively, the exclusive phosphorylation of ERK1/2 has been proposed to be a result of a possible tuning of the IGF-IR signaling cascade by the E-peptide toward MAPK (38).
The identification of the signaling pathways and effectors upstream and downstream of the E-peptide-induced activation of the extracellular-regulated kinases ERK1 and ERK2, the best-characterized members of the MAP kinase family (152), would provide information of particular importance regarding the mechanisms of E-peptide bioactivity. Furthermore, it is tempting to speculate that the selective activation of MAPK/ERK1/2 pathway up to a critical level by the E-peptide might further exert an inhibitory regulation of its competitive PI3K/Akt pathway, since it has been shown that the activity of one pathway might inactivate portions of the other (153,154), thus consisting of a regulatory mechanism of competitive biological actions such as cell proliferation and differentiation.
It should be also mentioned that, given the subcellular as well as differential localization of the E-peptides (94,141,155,156), an intracrine signaling mechanism that mediates E-peptides bioactivity, as proposed for some peptide growth factors (157), cannot be excluded.
Conclusion
IGF-I regulation occurs at multiple levels due to the alternative splicing of its pre-mRNA both at the 5′ and 3′ end, and the isoform-specific co- and posttranslational modifications, which appear to play key roles in modulating the bioavailabilty and bioactivity of this growth factor. New evidence supports the concept of IGF-I isoform- or IGF-I various peptides-specific functions, possibly via specific and divergent signaling. Nevertheless, it remains to be determined where the distinct E-peptide signaling diverges from that of mature IGF-I, or whether this signaling is induced uniquely and autonomously by the E-peptides or synergistically with the mature IGF-I bioactive molecule, and in the form of processed or pro-IGF-I molecules. Thus, the regulatory mechanism(s) of a potentially specific and apportioned bioactivity of the various IGF-I peptides is of particular interest within the context of a revisited characterization of the multiple IGF-I actions.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. (1993) Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell. 75:59–72.
Laviola L, Natalicchio A, Giorgino F. (2007) The IGF-I signaling pathway. Curr. Pharm. Des. 13:663–9.
Barton ER, et al. (2012) Deletion of muscle GRP94 impairs both muscle and body growth by inhibiting local IGF production. FASEB J. 26:3691–702.
Jones JI, Clemmons DR. (1995) Insulin-like growth factors and their binding proteins: biological actions. Endocr. Rev. 16:3–34.
Koutsilieris M, Mitsiades C, Sourla A. (2000) Insulin-like growth factor I and urokinase-type plasminogen activator bioregulation system as a survival mechanism of prostate cancer cells in osteoblastic metastases: development of anti-survival factor therapy for hormone-refractory prostate cancer. Mol. Med. 6:251–67.
Siddle K, et al. (2001) Specificity in ligand binding and intracellular signalling by insulin and insulin-like growth factor receptors. Biochem. Soc. Trans. 29:513–25.
LeRoith D, Roberts CT, Jr. (2003) The insulin-like growth factor system and cancer. Cancer Lett. 195:127–37.
Kooijman R. (2006) Regulation of apoptosis by insulin-like growth factor (IGF)-I. Cytokine Growth Factor Rev. 17:305–23.
Werner H, LeRoith D. (1996) The role of the insulin-like growth factor system in human cancer. Adv. Cancer Res. 68:183–223.
Reyes-Moreno C, Sourla A, Choki I, Doillon C, Koutsilieris M. (1998) Osteoblast-derived survival factors protect PC-3 human prostate cancer cells from adriamycin apoptosis. Urology. 52:341–7.
Werner H, Bruchim I. (2009) The insulin-like growth factor-I receptor as an oncogene. Arch. Physiol. Biochem. 115:58–71.
Ewton DZ, Roof SL, Magri KA, McWade FJ, Florini JR. (1994) IGF-II is more active than IGF-I in stimulating L6A1 myogenesis: greater mitogenic actions of IGF-I delay differentiation. J. Cell Physiol. 161:277–84.
Florini JR, Ewton DZ, Coolican SA. (1996) Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 17:481–517.
Daughaday WH, Rotwein P. (1989) Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr. Rev. 10:68–91.
Ohlsson C, et al. (2009) The role of liver-derived insulin-like growth factor-I. Endocr. Rev. 30:494–535.
Naranjo WM, et al. (2002) Protein calorie restriction affects nonhepatic IGF-I production and the lymphoid system: studies using the liver-specific IGF-I gene-deleted mouse model. Endocrinology. 143:2233–41.
Klover P, Hennighausen L. (2007) Postnatal body growth is dependent on the transcription factors signal transducers and activators of transcription 5a/b in muscle: a role for autocrine/paracrine insulin-like growth factor I. Endocrinology. 148:1489–97.
Stratikopoulos E, Szabolcs M, Dragatsis I, Klinakis A, Efstratiadis A. (2008) The hormonal action of IGF1 in postnatal mouse growth. Proc. Natl. Acad. Sci. U. S. A. 105:19378–83.
Duan C, Ren H, Gao S. (2010) Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen. Comp. Endocrinol. 167:344–51.
Siegfried JM, et al. (1992) A mitogenic peptide amide encoded within the E peptide domain of the insulin-like growth factor IB prohormone. Proc. Natl. Acad. Sci. U. S. A. 89:8107–11.
Gilmour RS. (1994) The implications of insulinlike growth factor mRNA heterogeneity. J. Endocrinol. 140:1–3.
Chew SL, Lavender P, Clark AJ, Ross RJ. (1995) An alternatively spliced human insulin-like growth factor-I transcript with hepatic tissue expression that diverts away from the mitogenic IBE1 peptide. Endocrinology. 136:1939–44.
Wallis M. (2009) New insulin-like growth factor (IGF)-precursor sequences from mammalian genomes: the molecular evolution of IGFs and associated peptides in primates. Growth Horm. IGF. Res. 19:12–23.
Rotwein P, Pollock KM, Didier DK, Krivi GG. (1986) Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J. Biol. Chem. 261:4828–32.
Duguay SJ, Lai-Zhang J, Steiner DF. (1995) Mutational analysis of the insulin-like growth factor I prohormone processing site. J. Biol. Chem. 270:17566–74.
Barton ER. (2006) The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl. Physiol. Nutr. Metab. 31:791–7.
Kuo YH, Chen TT. (2002) Novel activities of pro IGF-I E peptides: regulation of morphological differentiation and anchorage-independent growth in human neuroblastoma cells. Exp. Cell Res. 280:75–89.
Mills P, Dominique JC, Lafreniere JF, Bouchentouf M, Tremblay JP. (2007) A synthetic mechano growth factor E Peptide enhances myogenic precursor cell transplantation success. Am. J. Transplant. 7:2247–59.
Dluzniewska J, et al. (2005) A strong neuroprotective effect of the autonomous C-terminal peptide of IGF-1 Ec (MGF) in brain ischemia. FASEB J. 19:1896–8.
Pfeffer LA, Brisson BK, Lei H, Barton ER. (2009) The insulin-like growth factor (IGF)-I E-peptides modulate cell entry of the mature IGF-I protein. Mol. Biol. Cell. 20:3810–7.
Philippou A, et al. (2009) Expression of IGF-1 isoforms after exercise-induced muscle damage in humans: characterization of the MGF E peptide actions in vitro. In Vivo. 23:567–75.
Quesada A, Ogi J, Schultz J, Handforth A. (2011) C-terminal mechano-growth factor induces heme oxygenase-1-mediated neuroprotection of SH-SY5Y cells via the protein kinase C/Nrf2 pathway. J. Neurosci. Res. 89:394–405.
Stavropoulou A, et al. (2009) IGF-1 expression in infarcted myocardium and MGF E peptide actions in rat cardiomyocytes in vitro. Mol. Med. 15:127–35.
Armakolas A, et al. (2010) Preferential expression of IGF-1Ec (MGF) transcript in cancerous tissues of human prostate: evidence for a novel and autonomous growth factor activity of MGF E peptide in human prostate cancer cells. Prostate. 70:1233–42.
Milingos DS, et al. (2010) Insulinlike growth factor-1Ec (MGF) expression in eutopic and ectopic endometrium: characterization of the MGF E-peptide actions in vitro. Mol. Med. 17:21–8.
Barton ER, DeMeo J, Lei H. (2010) The insulinlike growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J. Appl. Physiol. (1985). 108:1069–76.
Gentile MA, et al. (2010) Androgen-mediated improvement of body composition and muscle function involves a novel early transcriptional program including IGF1, mechano growth factor, and induction of ta-catenin. J. Mol. Endocrinol. 44:55–73.
Brisson BK, Barton ER. (2012) Insulin-like growth factor-I E-peptide activity is dependent on the IGF-I receptor. PLoS One. 7:e45588.
Quesada A, Micevych P, Handforth A. (2009) C-terminal mechano growth factor protects dopamine neurons: a novel peptide that induces heme oxygenase-1. Exp. Neurol. 220:255–66.
Deng M, et al. (2011) Mechano growth factor E peptide promotes osteoblasts proliferation and bone-defect healing in rabbits. Int. Orthop. 35:1099–106.
Adamo ML, et al. (1993) Distinct promoters in the rat insulin-like growth factor-I (IGF-I) gene are active in CHO cells. Endocrinology. 132:935–937.
Yang H, et al. (1995) Alternative leader sequences in insulin-like growth factor I mRNAs modulate translational efficiency and encode multiple signal peptides. Mol. Endocrinol. 9:1380–95.
Shavlakadze T, Winn N, Rosenthal N, Grounds MD. (2005) Reconciling data from transgenic mice that overexpress IGF-I specifically in skeletal muscle. Growth Horm. IGF. Res. 15:4–18.
Philippou A, Maridaki M, Halapas A, Koutsilieris M. (2007) The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In Vivo. 21:45–54.
Okazaki R, Durham SK, Riggs BL, Conover CA. (1995) Transforming growth factor-beta and forskolin increase all classes of insulin-like growth factor-I transcripts in normal human osteoblast-like cells. Biochem. Biophys. Res. Commun. 207:963–70.
Bloor CA, Knight RA, Kedia RK, Spiteri MA, Allen JT. (2001) Differential mRNA expression of insulin-like growth factor-1 splice variants in patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 164:265–72.
Shamblott MJ, Chen TT. (1993) Age-related and tissue-specific levels of five forms of insulin-like growth factor mRNA in a teleost. Mol. Mar. Biol. Biotechnol. 2:351–61.
Duguay SJ, Swanson P, Dickhoff WW. (1994) Differential expression and hormonal regulation of alternatively spliced IGF-I mRNA transcripts in salmon. J Mol. Endocrinol. 12:25–37.
Faulk CK, Perez-Dominguez R, Webb KA, Jr., Holt GJ. (2010) The novel finding of four distinct prepro-IGF-I E domains in a perciform fish, Sciaenops ocellatus, during ontogeny. Gen. Comp. Endocrinol. 169:75–81.
Zhang J, et al. (2013) Cloning and characterization of new transcript variants of insulin-like growth factor-I in Sika deer (Cervus elaphus). Growth Horm. IGF. Res. 23:120–7.
Jansen E, Steenbergh PH, LeRoith D, Roberts CT, Jr., Sussenbach JS. (1991) Identification of multiple transcription start sites in the human insulin-like growth factor-I gene. Mol. Cell. Endocrinol. 78:115–25.
Philippou A, Armakolas A, Koutsilieris M. (2013) Evidence for the Possible Biological Significance of the igf-1 Gene Alternative Splicing in Prostate Cancer. Front Endocrinol. (Lausanne). 4:31.
Adamo ML, Ben-Hur H, LeRoith D, Roberts CT, Jr. (1991) Transcription initiation in the two leader exons of the rat IGF-I gene occurs from disperse versus localized sites. Biochem. Biophys. Res. Commun. 176:887–93.
D’Ercole AJ, Stiles AD, Underwood LE. (1984) Tissue concentrations of somatomedin C: further evidence for multiple sites of synthesis and paracrine or autocrine mechanisms of action. Proc. Natl. Acad. Sci. U. S. A. 81:935–9.
Rotwein P, Pollock KM, Watson M, Milbrandt JD. (1987) Insulin-like growth factor gene expression during rat embryonic development. Endocrinology. 121:2141–4.
Adamo M, Lowe WL, Jr., LeRoith D, Roberts CT, Jr. (1989) Insulin-like growth factor I messenger ribonucleic acids with alternative 5′-untranslated regions are differentially expressed during development of the rat. Endocrinology. 124:2737–44.
Wang X, Yang Y, Adamo ML. (1997) Characterization of the rat insulin-like growth factor I gene promoters and identification of a minimal exon 2 promoter. Endocrinology. 138:1528–36.
O’Sullivan DC, Szestak TA, Pell JM. (2002) Regulation of IGF-I mRNA by GH: putative functions for class 1 and 2 message. Am. J. Physiol. Endocrinol. Metab. 283:E251–8.
Woelfle J, Chia DJ, Rotwein P. (2003) Mechanisms of growth hormone (GH) action. Identification of conserved Stat5 binding sites that mediate GH-induced insulin-like growth factor-I gene activation. J. Biol. Chem. 278:51261–6.
Chia DJ, Young JJ, Mertens AR, Rotwein P. (2010) Distinct alterations in chromatin organization of the two IGF-I promoters precede growth hormone-induced activation of IGF-I gene transcription. Mol. Endocrinol. 24:779–89.
Lund PK. (1994) Insulin-like growth factor I: molecular biology and relevance to tissue-specific expression and action. Recent Prog. Horm. Res. 49:125–48.
Hameed M, et al. (2004) The effect of recombinant human growth hormone and resistance training on IGF-I mRNA expression in the muscles of elderly men. J. Physiol. 555:231–40.
Imanaka M, et al. (2008) Growth hormone stimulates mechano growth factor expression and activates myoblast transformation in C2C12 cells. Kobe J. Med. Sci. 54:E46–54.
Aperghis M, et al. (2009) Serum IGF-I levels and IGF-I gene splicing in muscle of healthy young males receiving rhGH. Growth Horm. IGF. Res. 19:61–7.
Moschos MM, et al. (2011) Expression of the insulin-like growth factor 1 (IGF-1) and type I IGF receptor mRNAs in human HLE-B3 lens epithelial cells. In Vivo. 25:179–84.
Jansen M, et al. (1983) Sequence of cDNA encoding human insulin-like growth factor I precursor. Nature. 306:609–11.
Rotwein P. (1986) Two insulin-like growth factor I messenger RNAs are expressed in human liver. Proc. Natl. Acad. Sci. U. S. A. 83:77–81.
McKay BR, O’Reilly CE, Phillips SM, Tarnopolsky MA, Parise G. (2008) Co-expression of IGF-1 family members with myogenic regulatory factors following acute damaging muscle-lengthening contractions in humans. J. Physiol. 586:5549–60.
Koczorowska MM, Kwasniewska A, Gozdzicka-Jozefiak A. (2011) IGF1 mRNA isoform expression in the cervix of HPV-positive women with pre-cancerous and cancer lesions. Exp. Ther. Med. 2:149–56.
Shimatsu A, Rotwein P. (1987) Sequence of two rat insulin-like growth factor I mRNAs differing within the 5′ untranslated region. Nucleic Acids Res. 15:7196.
Goldspink G. (2005) Mechanical signals, IGF-I gene splicing, and muscle adaptation. Physiology (Bethesda). 20:232–8.
Matheny RW, Jr., Nindl BC, Adamo ML. (2010) Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology. 151:865–875.
Philippou A, et al. (2011) IGF1Ec expression in MG-63 human osteoblast-like osteosarcoma cells. Anticancer Res. 31:4259–65.
Yu H, Rohan T. (2000) Role of the insulin-like growth factor family in cancer development and progression. J. Natl. Cancer Inst. 92:1472–89.
Mourkioti F, Rosenthal N. (2005) IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 26:535–42.
Temmerman L, Slonimsky E, Rosenthal N. (2010) Class 2 IGF-1 isoforms are dispensable for viability, growth and maintenance of IGF-1 serum levels. Growth Horm. IGF. Res. 20:255–63.
Zhang J, Whitehead RE, Jr., Underwood LE. (1997) Effect of fasting on insulin-like growth factor (IGF)-IA and IGF-IB messenger ribonucleic acids and prehormones in rat liver. Endocrinology. 138:3112–8.
Duguay SJ. (1999) Post-translational processing of insulin-like growth factors. Horm. Metab. Res. 31:43–9.
Powell DR, Lee PD, Chang D, Liu F, Hintz RL. (1987) Antiserum developed for the E peptide region of insulin-like growth factor IA prohormone recognizes a serum protein by both immunoblot and radioimmunoassay. J. Clin. Endocrinol. Metab 65:868–75.
Wilson HE, Westwood M, White A, Clayton PE. (2001) Monoclonal antibodies to the carboxy-terminal Ea sequence of pro-insulin-like growth factor-IA (proIGF-IA) recognize proIGF-IA secreted by IM9 B-lymphocytes. Growth Horm. IGF. Res. 11:10–7.
Kravchenko IV, Furalyov VA, Khotchenkov VP, Popov VO. (2006) Monoclonal antibodies to mechano-growth factor. Hybridoma (Larchmt). 25:300–5.
Philippou A, et al. (2008) Characterization of a rabbit antihuman mechano growth factor (MGF) polyclonal antibody against the last 24 amino acids of the E domain. In Vivo. 22:27–35.
Lingappa VR, Lingappa JR, Blobel G. (1980) Signal sequences for early events in protein secretion and membrane assembly. Ann. N. Y. Acad. Sci. 343:356–61.
Denley A, Cosgrove LJ, Booker GW, Wallace JC, Forbes BE. (2005) Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 16:421–39.
Carlsson-Skwirut C, et al. (1986) Isolation and characterization of variant IGF-1 as well as IGF-2 from adult human brain. FEBS Lett 201:46–50.
Sara VR, et al. (1993) The biological role of truncated insulin-like growth factor-1 and the tripeptide GPE in the central nervous system. Ann. N. Y. Acad. Sci. 692:183–91.
Nakayama K. (1997) Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327 (Pt 3): 625–35.
Duckert P, Brunak S, Blom N. (2004) Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17:107–12.
Rouille Y, et al. (1995) Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol. 16:322–61.
Steiner DF. (1998) The proprotein convertases. Curr. Opin. Chem. Biol. 2:31–9.
Seidah NG, Chretien M. (1999) Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848:45–62.
Duguay SJ, Milewski WM, Young BD, Nakayama K, Steiner DF. (1997) Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J. Biol. Chem. 272:6663–70.
Thomas G. (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 3:753–66.
Tan NS, Ho B, Ding JL. (2002) Engineering a novel secretion signal for cross-host recombinant protein expression. Protein Eng. 15:337–45.
Conover CA, Baker BK, Hintz RL. (1989) Cultured human fibroblasts secrete insulin-like growth factor IA prohormone. J. Clin. Endocrinol. Metab. 69:25–30.
Conover CA, et al. (1993) Human hepatoma cells synthesize and secrete insulin-like growth factor Ia prohormone under growth hormone control. Regul. Pept. 48:1–8.
Durzynska J, Philippou A, Brisson BK, Nguyen-McCarty M, Barton ER. (2013) The pro-forms of insulin-like growth factor I (IGF-I) are predominant in skeletal muscle and alter IGF-I receptor activation. Endocrinology. 154:1215–24.
Tsuji A, et al. (2003) Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim. Biophys. Acta. 1645:95–104.
Marshall RD. (1972) Glycoproteins. Annu. Rev. Biochem. 41:673–702.
Imperiali B, O’Connor SE, Hendrickson T, Kellenberger C. (1999) Chemistry and biology of asparagine-linked glycosylation. Pure Appl. Chem. 71:777–87.
Simons G, et al. (1993) Overproduction of bovine beta-casein in Escherichia coli and engineering of its main chymosin cleavage site. Protein Eng. 6:763–70.
Freeze HH. (2001) Endoglycosidase and glycoamidase release of N-linked oligosaccharides. Curr. Protoc. Mol. Biol. Chapter 17:Unit17 13A.
Hede MS, et al. (2012) E-peptides control bioavailability of IGF-1. PLoS One. 7:e51152.
Metzger F, et al. (2011) Separation of fast from slow anabolism by site-specific PEGylation of insulin-like growth factor I (IGF-I). J. Biol. Chem. 286:19501–10.
Sivaramakrishnan M, et al. (2013) PEGylation of lysine residues reduces the pro-migratory activity of IGF-I. Biochim. Biophys. Acta. 1830:4734–42.
Rinderknecht E, Humbel RE. (1978) The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J. Biol. Chem. 253:2769–76.
Federici M, et al. (1997) Increased abundance of insulin/IGF-I hybrid receptors in adipose tissue from NIDDM patients. Mol. Cell. Endocrinol. 135:41–7.
Nakae J, Kido Y, Accili D. (2001) Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 22:818–35.
Taguchi A, White MF. (2008) Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 70:191–212.
De Meyts P, Whittaker J. (2002) Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug. Discov. 1:769–83.
Kristensen C, Wiberg FC, Andersen AS. (1999) Specificity of insulin and insulin-like growth factor I receptors investigated using chimeric mini-receptors. Role of C-terminal of receptor alpha subunit. J. Biol. Chem. 274:37351–6.
Baserga R, Peruzzi F, Reiss K. (2003) The IGF-1 receptor in cancer biology. Int. J. Cancer 107:873–7.
Manes S, et al. (1997) Functional epitope mapping of insulin-like growth factor I (IGF-I) by anti-IGF-I monoclonal antibodies. Endocrinology. 138:905–15.
Philippou A, Halapas A, Maridaki M, Koutsilieris M. (2007) Type I insulin-like growth factor receptor signaling in skeletal muscle regeneration and hypertrophy. J. Musculoskelet. Neuronal Interact. 7:208–18.
Wu J, et al. (2008) Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor. EMBO. J. 27:1985–94.
Ozkan EE. (2011) Plasma and tissue insulin-like growth factor-I receptor (IGF-IR) as a prognostic marker for prostate cancer and anti-IGF-IR agents as novel therapeutic strategy for refractory cases: a review. Mol. Cell. Endocrinol. 344:1–24.
Hubbard SR. (1997) Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16:5572–81.
Baserga R. (1999) The IGF-I receptor in cancer research. Exp. Cell. Res. 253:1–6.
Baxter RC. (2000) Insulin-like growth factor (IGF)-binding proteins: interactions with IGFs and intrinsic bioactivities. Am. J. Physiol. Endocrinol. Metab. 278:E967–76.
Duan C. (2002) Specifying the cellular responses to IGF signals: roles of IGF-binding proteins. J. Endocrinol. 175:41–54.
Cohen P. (2006) Overview of the IGF-I system. Horm. Res. 65 Suppl 1:3–8.
Baxter RC, Martin JL, Beniac VA. (1989) High molecular weight insulin-like growth factor binding protein complex. Purification and properties of the acid-labile subunit from human serum. J. Biol. Chem. 264:11843–8.
Baxter RC, Martin JL. (1989) Structure of the Mr 140,000 growth hormone-dependent insulin-like growth factor binding protein complex: determination by reconstitution and affinity-labeling. Proc. Natl. Acad. Sci. U. S. A. 86:6898–902.
Kelley KM, et al. (1996) Insulin-like growth factor-binding proteins (IGFBPs) and their regulatory dynamics. Int. J. Biochem. Cell Biol. 28:619–37.
Clemmons DR. (1997) Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev. 8:45–62.
Firth SM, Baxter RC. (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23:824–54.
Oh Y. (1998) IGF-independent regulation of breast cancer growth by IGF binding proteins. Breast Cancer Res. Treat. 47:283–93.
Koutsilieris M, Lavergne E, Lemay A. (1997) Association of protease activity against IGFBP-3 with peritoneal fluid mitogens: possible implications for the ectopic growth of endometrial cells in women with endometriosis. Anticancer Res. 17:1239–44.
Hwa V, Oh Y, Rosenfeld RG. (1999) The insulinlike growth factor-binding protein (IGFBP) superfamily. Endocr. Rev. 20:761–87.
Hwa V, Oh Y, Rosenfeld RG. (1999) Insulin-like growth factor binding proteins: a proposed superfamily. Acta. Paediatr. Suppl. 88:37–45.
Lopez-Bermejo A, et al. (2006) Insulin resistance is associated with increased serum concentration of IGF-binding protein-related protein 1 (IGFBP-rP1/MAC25). Diabetes. 55:2333–9.
Barton ER. (2006) Viral expression of insulinlike growth factor-I isoforms promotes different responses in skeletal muscle. J. Appl. Physiol. (1985) 100:1778–84.
Carpenter V, et al. (2008) Mechano-growth factor reduces loss of cardiac function in acute myocardial infarction. Heart Lung Circ. 17:33–9.
Rae FK, et al. (2012) Proximal tubule overexpression of a locally acting IGF isoform, Igf-1Ea, increases inflammation after ischemic injury. Growth Horm. IGF. Res. 22:6–16.
Ates K, et al. (2007) The IGF-I splice variant MGF increases progenitor cells in ALS, dystrophic, and normal muscle. FEBS Lett 581:2727–32.
Poudel B, et al. (2011) Increased cardiogenesis in P19-GFP teratocarcinoma cells expressing the propeptide IGF-1Ea. Biochem. Biophys. Res. Commun. 416:293–9.
Kasprzak A, et al. (2012) Differential expression of IGF-1 mRNA isoforms in colorectal carcinoma and normal colon tissue. Int. J. Oncol. 42:305–16.
Vinciguerra M, Musaro A, Rosenthal N. (2010) Regulation of muscle atrophy in aging and disease. Adv. Exp. Med. Biol. 694:211–33.
Vinciguerra M, et al. (2012) mIGF-1/JNK1/SirT1 signaling confers protection against oxidative stress in the heart. Aging Cell. 11:139–49.
Santini MP, et al. (2007) Enhancing repair of the mammalian heart. Circ. Res. 100:1732–40.
Durzynska J, Wardzinski A, Koczorowska M, Gozdzicka-Jozefiak A, Barton ER. (2013) Human Eb peptide: not just a by-product of pre-pro-IGF1b processing? Horm. Metab. Res. 45:415–22.
Deng M, et al. (2012) New proangiogenic activity on vascular endothelial cells for C-terminal mechano growth factor. Acta. Biochim. Biophys. Sin. (Shanghai). 44:316–22.
Collins JM, Goldspink PH, Russell B. (2010) Migration and proliferation of human mesenchymal stem cells is stimulated by different regions of the mechano-growth factor prohormone. J. Mol. Cell Cardiol. 49:1042–5.
Chen MJ, et al. (2007) Suppression of growth and cancer-induced angiogenesis of aggressive human breast cancer cells (MDA-MB-231) on the chorioallantoic membrane of developing chicken embryos by E-peptide of pro-IGF-I. J. Cell. Biochem. 101:1316–27.
Yang SY, Goldspink G. (2002) Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett 522:156–60.
Qin LL, et al. (2013) Mechano growth factor (MGF) promotes proliferation and inhibits differentiation of porcine satellite cells (PSCs) by down-regulation of key myogenic transcriptional factors. Mol. Cell. Biochem. 370:221–30.
Tian XC, Chen MJ, Pantschenko AG, Yang TJ, Chen TT. (1999) Recombinant E-peptides of pro-IGF-I have mitogenic activity. Endocrinology. 140:3387–90.
Kuo YH, Chen TT. (2003) Specific cell surface binding sites shared by human Pro-IGF-I Eb-peptides and rainbow trout Pro-IGF-I Ea-4-peptide. Gen. Comp. Endocrinol. 132:231–40.
Matheny RW, Jr., Nindl BC. (2011) Loss of IGF-IEa or IGF-IEb impairs myogenic differentiation. Endocrinology. 152:1923–34.
Brisson BK, Barton ER. (2013) New Modulators for IGF-I Activity within IGF-I Processing Products. Front Endocrinol. (Lausanne) 4:42.
Malbon CC, Karoor V. (1998) G-protein-linked receptors as tyrosine kinase substrates: new paradigms in signal integration. Cell Signal. 10:523–7.
Blenis J. (1993) Signal transduction via the MAP kinases: proceed at your own RSK. Proc. Natl. Acad. Sci. U. S. A. 90:5889–92.
Samuel DS, et al. (1999) Raf-1 activation stimulates proliferation and inhibits IGF-stimulated differentiation in L6A1 myoblasts. Horm. Metab. Res. 31:55–64.
Rommel C, et al. (1999) Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 286:1738–41.
Zhang B, Xian C, Luo Y, Wang Y. (2009) Expression and subcellular localization of mechano-growth factor in osteoblasts under mechanical stretch. Sci. China C. Life Sci. 52:928–34.
Peng Q, et al. (2012) The nuclear localization of MGF receptor in osteoblasts under mechanical stimulation. Mol. Cell Biochem. 369:147–56.
Re RN. (2003) The intracrine hypothesis and intracellular peptide hormone action. Bioessays. 25:401–9.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (https://doi.org/creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
Philippou, A., Maridaki, M., Pneumaticos, S. et al. The Complexity of the IGF1 Gene Splicing, Posttranslational Modification and Bioactivity. Mol Med 20, 202–214 (2014). https://doi.org/10.2119/molmed.2014.00011
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2014.00011