Abstract
Chronic lymphocytic leukemia (CLL) cells survive longer in vivo than in vitro, suggesting that the tissue microenvironment provides prosurvival signals to tumor cells. Primary and secondary lymphoid tissues are involved in the pathogenesis of CLL, and the role of these tissue microenvironments has not been explored completely. To elucidate host-tumor interactions, we performed gene expression profiling (GEP) of purified CLL cells from peripheral blood (PB; n = 20), bone marrow (BM; n = 18), and lymph node (LN; n = 15) and validated key pathway genes by real-time polymerase chain reaction, immunohistochemistry and/or TCL1 trans-genic mice. Gene signatures representing several pathways critical for survival and activation of B cells were altered in CLL cells from different tissue compartments. Molecules associated with the B-cell receptor (BCR), B cell-activating factor/a proliferation-inducing ligand (BAFF/APRIL), nuclear factor (NF)-κB pathway and immune suppression signature were enriched in LN-CLL, suggesting LNs as the primary site for tumor growth. Immune suppression genes may help LN-CLL cells to modulate antigen-presenting and T-cell behavior to suppress antitumor activity. PB CLL cells overexpressed chemokine receptors, and their cognate ligands were enriched in LN and BM, suggesting that a chemokine gradient instructs B cells to migrate toward LN or BM. Of several chemokine ligands, the expression of CCL3 was associated with poor prognostic factors. The BM gene signature was enriched with antiapoptotic, cytoskeleton and adhesion molecules. Interestingly, PB cells from lymphadenopathy patients shared GEP with LN cells. In Eμ-TCL1 transgenic mice (the mouse model of the disease), a high percentage of leukemic cells from the lymphoid compartment express key BCR and NF-κB molecules. Together, our findings demonstrate that the lymphoid microenvironment promotes survival, proliferation and progression of CLL cells via chronic activation of BCR, BAFF/APRIL and NF-κB activation while suppressing the immune response.
Similar content being viewed by others
Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the Western world. CLL is a disease of long-lived B cells that are capable of proliferating upon appropriate stimuli and accumulate in the peripheral blood (PB), bone marrow (BM) and lymph nodes (LNs) (1–4). There is emerging evidence that the tumor microenvironment influences the survival and drug resistance of CLL cells (5) and other cancer cells (6,7), playing a critical role in the growth, invasion and progression of a variety of malignancies, including hematological malignancies. CLL cells rapidly undergo apoptosis in vitro, but survive for a longer time in vivo, thus underscoring the role of the microenvironment in the growth and survival of CLL cells (8). Several studies have used in vitro culture systems involving T cells, stromal cells, follicular dendritic cells, nurselike cells (NLCs) and CD40 engagement to study the role of the microenvironment in CLL (9–11). The presence of stromal components in the microenvironment, such as NLCs, protects CLL cells from death and enhances the expression of genes related to chemokines and B-cell receptor (BCR) signaling (9,12). Furthermore, CLL cells proliferate in vitro in the presence of stroma and soluble mediators such as interleukin (IL)-2 or IL-10 (13). Although these in vitro culture systems simulate the in vivo microenvironment to a certain extent, studying CLL cells directly from in vivo sites will more accurately define the molecules associated with vital functions in vivo.
Emerging evidence suggests that clinical/biological heterogeneity in CLL is due to tissue microenvironment-influenced variation in proliferation and survival (8). Only a few studies have reported the importance of PB, BM and LN microenvironments in CLL (14–19). This study enhances the information by providing a comprehensive list of gene signatures of CLL cells from three distinct tumor microenvironments, such as PB, BM and LN not reported previously, and associates these signatures to clinical markers that predict disease progression and clinical outcome.
Materials and Methods
CLL Patient Information and Sample Collection
PB (n = 20), BM (n = 18) and LN (n = 15) samples were collected from CLL patients with informed consent using an institutional review board-approved protocol at University of Nebraska Medical Center. Of these, seven PB and BM samples, two PB and LN, and three BM and LN paired samples were from the same patient. Only untreated patients and patients who had not received treatment in the prior 6 months were included in this study. The patient characteristics are described in Results.
Isolation and Characterization of CLL Cells
CLL cells from PB and BM were isolated and immunophenotyped by flow cytometry by using the following combinations of antibodies as described previously (20). Briefly, wherever needed, antibody cocktail for B-cell enrichment (RosetteSep; Stemcell Technologies Inc., Vancouver, BC, Canada) was used to purify the B-CLL cells from PB and BM samples. This cocktail kit binds and eliminates erythrocytes and other white blood cells except B cells by subsequent density gradient centrifugation using LymphoPrep and isolation of purified B cells (>90%) from the interface. Frozen LN samples were obtained from the University of Nebraska Medical Center tissue bank. CLL cells were identified in the 10-micron sections of these LNs by using immunohistochemical techniques to localize CD19+ and CD5+ cells. Fifteen to twenty (8–10 microns thick) sections were prepared from each LN on membrane-coated glass slides. These slides were prepared at 4°C, and these sections were immediately fixed with ice-cold acetone and stained with cresyl violet (LCM Staining Kit; Ambion/Life Technologies, Carlsbad, CA, USA) for 30 s. The corresponding CD19+ and CD5+ areas were laser microdissected by using the laser capture microdissection technique (Leica Microsystems Inc., Wetzlar, Germany), specially focusing on areas with >90% CLL cells, which were identified and microdissected. Areas containing stromal cells either were excluded or burnt out by using a laser, thus further decreasing the stromal cell components. The microdis-sected cells were immediately processed for isolation of RNA to avoid degradation of RNA.
Cytogenetic Analyses
The cytogenetic analyses including fluorescent in situ hybridization (FISH) were performed by the Human Genetics Institute at University of Nebraska Medical Center as described earlier (21). Chromosome 11q deletion, 17p deletion and tri-somy 12 were considered as the poor outcome group, whereas normal karyotype and 13q deletion were grouped as the better outcome group (22).
In Vitro Stromal Cell Culture System
To simulate the in vivo microenvironment, an in vitro stromal culture system was used to study the survival and proliferation of CLL cells in vitro as described earlier (25). Freshly isolated primary CLL cells were cocultured on the mouse-derived OMA-AD or human-derived HMEC stromal feeder cell layer in the presence of RPMI with 10% fetal bovine serum medium for 4872 h, and survival and proliferation of CLL cells were determined by flow cy-tometry.
Gene Expression Analyses
Total RNA was extracted from CLL cells by using TRIzol (Invitrogen/Life Technologies) as described earlier (20). RNAs (approximately 0.5 μg per sample) from CLL cells were used for gene expression profiling on a DNA microarray chip (MWG Biotech, Ebersberg, Germany, Human 30K oligo set B) consisting of 50-mer oligonucleotide representing 10,000 different genes. Stratagene reference RNA, labeling of cDNA, hybridization procedure and locally weighted scatterplot smoothing (LOWESS) intensity-dependent normalization were implemented using standard procedures, Gene Pix 6.0 and BRB Array Tools as described previously (20). Differential mRNA expression in PB-, BM- and LN-CLL cells was evaluated by using a random variance t test (p < 0.005), significance analysis of microarrays (false discovery rate [FDR] <10%) and gene set enrichment analysis computational program in conjunction with BRB array tools (version 4.2.0-Beta) (23,24). Cluster and Tree View programs were also used in the analyses (Eisen Laboratory, University of California, Berkeley, CA, USA).
Validation of Significant Genes Using Real-Time Quantitative Polymerase Chain Reaction
SYBR Green real-time polymerase chain reaction (PCR) was used to further confirm differential gene expression between CLL groups. Complementary cDNAs were mixed with primers and Power SYBR Green PCR Master Mix (Applied Biosystems/Life Technologies) as previously described (25).
Detection of Surface and Intracellular Markers Using Flow Cytometry
Cells were stained with CD19-FITC (fluorescein isothiocyanate) marker to specifically analyze the proportion of CLL cells. Further, surface apoptotic marker annexin V, intracellular phospho-Syk and proliferation marker Ki-67 were detected by using annexin V/PI staining, “phosphoflow” (BD Phosflow) and Ki-67 staining, respectively, following the manufacturer protocol (BD Biosciences, San Jose, CA, USA). For analyses, a BD FAC-Star Plus flow cytometer (BD Biosciences) was used.
Identification of Key Signaling Molecules Using Eμ-TCL1 Transgenic Mouse
TCL1 transgenic (TCL1-tg, n = 3), a mouse model for CLL, and C57BL/6 control (n = 3) mice were a generous gift from our collaborator Rene Opavsky at University of Nebraska Medical Center. These mice were reared and maintained at a pathogen-free animal facility in the University of Nebraska Medical Center. LN and spleen tissues were harvested from these mice to study key molecules in the leukemic cells. All experiments were performed in accord with the approved protocol by the Institutional Animal Care and Use Committee of University of Nebraska Medical Center.
Evaluation of Expression of Key Molecules at the Protein Level Using Immunohistochemistry
To validate our findings from the gene expression analyses, we examined the expression levels of phosphor-SYK and phosphor-p-65, the major molecules associated with BCR and nuclear factor (NF)-κB signaling using immunochem-istry. Briefly, the 5- to 10-micron histolog-ical sections of the LNs from CLL patients and lymphoid tissue from TCL1 transgenic mice were stained with p-SYK (catalog number PAB0623, 1:200 dilution) or p-P65 (catalog number AB28810, 1:50 dilution). A Leica Bond Polymer Refine Detection Kit (Leica Microsystems Inc.) was used to visualize the cells positive for these two primary antibodies, as described in the manufacturer protocols.
Statistical Analyses
For the identification of differentially expressed genes, a significant analysis of microarray was used. To identify the tissue-specific gene signatures, analysis was performed using a random variance F test with a p value of 0.01 and FDR of 0.08. Most of the analyses were performed at p< 0.05 and FDR < 0.25, unless specified otherwise. The KaplanMeier method using the log-rank test was used to study the association of gene expression or clinical parameter with the clinical outcome as done previously (22,25). Time to treatment among CLL patients was used as an outcome and defined as the time period in months between diagnosis and initiation of the first treatment regimen.
All supplementary materials are available online at https://doi.org/www.molmed.org .
Results
Patient Characteristics
We performed GEP on 20 PB, 18 BM and 15 LN specimens from 37 consenting CLL patients. The clinical characteristics of these 37 patients are shown in Supplementary Table S1. The patients included 25 males (67.5%) with a median age of 55 years and 12 females (32.5%) with a median age of 65 years. The median age of the patients at the time of diagnosis was 59 years (range 29–85). Of the 37 patients, GEP was obtained from paired BM/PB (n = 7), BM/LN (n = 3) and LN/PB (n = 2) samples from the same patient. We also tested an additional cohort of 40 PB samples (20) from CLL patients to validate the biologic and clinical findings.
Mouse- and Human-Derived Stroma Supports CLL Cell Growth
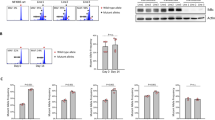
To investigate the influence of the microenvironments on the survival and growth of CLL cells, we used an in vitro system and cocultured CLL cells with stromal cells using BM-derived (OMA-AD) and endothelial-derived (HMEC) stromal cell lines (Figure 1A). Consistent with our previous observation, cocultured CLL cells showed a tendency of higher proliferation, as measured by a 1.5- and 3.5-fold increase in Ki-67–positive CLL cells, in OMA-AD and HMEC cocultured conditions, respectively. Consistent with this observation, prolonged survival was observed with significantly (p < 0.0001, p < 0.005) reduced frequency of early apoptotic (positive for annexin V only) and late apoptotic (positive for annexin V/PI) co-cultured CLL cells under both types of stromal cell systems (Figure 1B). Interestingly, cocultured CLL cells demonstrated significant (p < 0.05) upregulation of NF-κB target genes, for example, IL-6, BCL2 and TRAF1, as measured by quantitative real-time (qRT)-PCR (Figure 1). Thus, these studies indicate the importance of a compact stromal microenvironment usually found in tissues such as LNs in providing prosurvival and proproliferation signals to CLL cells.
Survival, proliferation and upregulation of the NF-κB-associated genes in cocultured CLL cells in vitro with human- or mouse-derived stromal cells. Analyses of cell survival, proliferation and change in expression of key genes in primary CLL cells cocultured with mouse BM-derived OMA-AD and human endothelial-derived HMEC stromal cells at 72 h. The results were analyzed using a Student t test (p < 0.05). (A) Measurement of the proliferation of CLL cells (n = 6) cocultured on stroma using Ki-67 staining. (B) Measurement of the survival of CLL cells (n < 7) cocultured on stroma using annexin V staining. (C) Validation of the upregulation of key genes in CLL cells (n = 6) cocultured on stroma using real-time PCR.
Gene Expression Signature of PB-, BM- and LN-CLL Cells
In our initial analyses using unsuper-vised hierarchical clustering of ~9,900 genetic probes on GEP of CLL cells from three in vivo sites, we observed that CLL cells from the three anatomic sites did not form tight clusters, indicating heterogeneity within the tissue compartments (Figure 2A). However, of the samples from the three lymphoid compartments, LN specimens (n = 15) clustered into two major nodes, whereas PB-CLL and BM-CLL samples were interspersed randomly, suggesting more homogeneity of gene expression among LN-CLL cells compared with PB-CLL and BM-CLL samples. These two groups of LNs were not distinguished from each other based on the known chromosomal abnormalities, high versus low CD38 expression, presence or absence of lymphadenopathy and/or time to treatment. Therefore, to obtain tissue-specific CLL gene signatures, supervised differential gene expression analysis was performed using a random variance F test (p < 0.01, FDR < 0.08). With this approach, we observed ~1,000 genes that were differentially expressed among PB-, BM-, and LN-CLL cells (Figure 2B). A detailed analysis showed that LN-CLL cells overexpressed various gene signatures involved in B-cell activation (CD40, SYK, TACI, BCMA), chemokine receptor signaling (CXCL13, CXCL14, CCL21) and induction of immune tolerance (CAV1, BATF, TTK, INDO1, IL10, CAMLG) (Figure 2B, Supplementary Figures S1A, B). Further, genes associated with mitogen-activated protein kinase (MAPK)-, phosphatidyli-nositide 3-kinase/protein kinase B (PI3K/AKT)- and NF-KB–signaling pathways were distinctly expressed in CLL cells from PB, BM and/or LN sites. Additionally, transcripts involved in B-cell activation and differentiation (CXCR4, FCER2, AKT2) were noted in the PB signature, whereas cytoskeleton and cell ad hesion molecules (KRT19, KRT18, COL10A1) were noted in the BM-CLL signature (Figure 2B, Supplementary Fig ure S1).
Unsupervised and supervised clustering of gene expression profiles of PB-CLL (n = 20), BM-CLL (n = 18) and LN-CLL (n = 15) cells in vivo from patients. (A) Unsupervised hierarchical clustering of genes obtained from CLL enriched lymphoid compartments. (B) Supervised clustering of genes of CLL cells isolated from different lymphoid compartments (p < 0.01, FDR < 0.08).
Differentially Expressed Genes Associated with the Seven Major Signaling Pathways
We next analyzed gene expression of CLL cells from different sites by using gene set enrichment analysis (24) to identify enriched cellular pathways/signatures. We observed >100 enriched cellular pathways/signatures among the CLL cells from LN, BM and PB. These analyses showed that LN-CLL cells were enriched in gene signatures related to tumor progression, and immunosuppres-sion or immune tolerance, suggesting that the LN microenvironment is conducive to tumor cell growth while inhibiting host immune response against CLL (Table 1; Supplementary Figures S2A, B, D, E). Additionally, the poor prognostic IGHV3-21 gene signature (26) consisting of genes associated with a worse outcome in CLL was enriched in LN-CLL compared with PB-CLL (Table 1, Supplementary Figure S2F). BM-CLL showed enrichment of gene signatures involved with cell cycle and antiapo-ptotic compared with PB-CLL (Table 1, Supplementary Figure S2C).
From the several enriched pathways differentially expressed among PB-, BM-and LN-CLL cells, seven major pathways/functional signatures relevant to CLL and/or B-cell biology were identified based on gene ontology: BCR signaling, B cell-activating factor/a proliferation-inducing ligand (BAFF/APRIL) signaling, MAPK signaling, PI3K/AKT signaling, NF-κB signaling, chemokine ligands/receptors and tolerance (Figure 3A). Further, the expression of critical genes in these signatures was validated by qRT-PCR. Only genes with significant differential expression (t test, p < 0.05) are shown in Figure 3. Several transcripts involved in BCR signaling (for example, SYK, BTK and VAV2) and BAFF/APRIL signaling (for example, BAFF, BCMA and TRAF2) that were significantly expressed in LN-CLL by microarray were confirmed by qRT-PCR (Figure 3B, Supplementary Table S2), thus suggesting activated BCR and BAFF/APRIL signaling in LN compared with PB or BM. Despite the absence of probes for ZAP-70 on our mi-croarray, we studied its expression using real-time PCR and found that it was significantly (p < 0.05) overexpressed in LN-CLL compared with PB-CLL and BM-CLL (Figure 3B). Similarly, transcripts of different genes associated with NF-κB (IL6, NF-κBIL1, RELB, STATT), MAPK (MAP3K2, MAP2K5, MAP2K11, MAPK4) and PI3K/Akt (AKT1, 4E-BP1, PDK1, IGFBP6) pathways that were overex-pressed in CLL cells from the different microenvironments (Figure 2, Supplementary Figure S1) were validated by qRT-PCR (Figure 3B). These pathways play an active role in the proliferation of CLL cells. It is likely that CLL cells in different microenvironments use different molecules for survival/proliferation.
Mean expression of genes associated with seven major signaling pathways and validation of expression of key genes using real-time PCR. (A) Supervised cluster analyses of differentially expressed genes (p < 0.05) associated with seven major signaling pathways. The mean expressions of these significant genes are shown among PB, BM and LN cells. (B) Confirmation of the differentially expressed genes among PB-CLL, BM-CLL and LN-CLL using real-time PCR. Expression of the selected genes from each major pathway was studied by using real-time PCR. Significant differences in expression levels between PB-CLL, BM-CLL and LN-CLL were determined by using the Student t test (p values when comparing with LN cells: *p < 0.05, **p < 0.01, #p < 0.005, ##p < 0.001; p values for comparison with PB is denoted with *p and comparison with BM is denoted as *b). First row: BCR signaling: SYK, BTK, ZAP70; second row: BAFF/APRIL signaling: BAFF, BCMA, TRAF2; third row: MAPK signaling: MAP2K6, CAMLG, STAT1; fourth row: PI3K/Akt pathway: PDK1, IGFBP6, AKT1; fifth row: NF-κB pathway: NF-κBIB, FCER2, CCND2; sixth row: chemokine ligands/receptors: CXCR4, CCL21, CCR7; seventh row: tolerogenic signature: CAV1, MCM3, IDH; microarray analyses had shown all genes were differentially expressed among PB-CLL (n = 20), BM-CLL (n = 18) and LN-CLL (n = 15).
From the GEP data, we surmised that CLL cells use chemokine ligands/receptors for migration; for example, CXCR4 and CCR7, were significantly overexpressed in PB-CLL and their cognate ligands CXCL12 and CCL21 were overexpressed in BM and LN microenvironments (Figures 2B and 3). These data are consistent with chemokine receptor/ligand gradients governing migration of CLL cells into different microenvironments. Interestingly, genes associated with immune tolerogenesis (CAV1, MCM3, BATF) were uniquely overex-pressed in LN-CLL compared with BM-CLL and PB-CLL (Figures 2B and 3, Supplementary Figure S1), indicating the LN microenvironment plays a role in inducing immunological tolerance of CLL cells. The role of this tolerogenic signature in LN is recently validated by us (38). Together, these results elucidate the genes overexpressed in LN-CLL and indicate that the microenvironment influences CLL cell behavior. To further confirm the LN microenvironment-induced effects on CLL progression, the expression of previously reported drug resistance-related genes was studied (27,28). Among the genes we analyzed, ABCC6, HSP70 and HSP90 were significantly overexpressed in LN-CLL compared with BM-CLL and PB-CLL cells, indicating that the LN microenvironment also induces drug resistance in CLL (data not shown). We validated the expression of phosphor-SYK (BCR signaling) and p-P65 (NF-κB signaling) in leukemic cells from TCL1 transgenic mouse and patients’ LN by using immunohistochem-istry (Figure 4). LN tissue was harvested from TCL1-Tg mice at 37 wks, at which time accumulation of leukemic B-cells (B220/CD5+) occur in lymphoid organs (40). A high percentage of leukemic B cells from LN of TCL1-Tg mice were found to be positive for p-SYK (~50%) and p-P65 (~99%) (Figure 4, panel I: A–F). The expression pattern of p-SYK and p-65 in leukemic cells from spleen tissue of TCL1-Tg mice at 37 wks were highly similar with that of LN from the mice (data not shown). Furthermore, the expression of these key signaling molecules were investigated in patient LNs. As expected, a high percentage of CLL cells were positive for p-SYK (~90%) and p-P65 (100%) signaling molecules, validating the expression signature in LN-CLL cells (Figure 4, panel II: C, D, F). Altogether, CLL cells from a defined structural microenvironment such as LN appears to have activated molecules of signaling pathways that are known to drive cell survival and proliferation.
Confirmation of expression of pSyk and p-P65 in lymphoid tissue of a TCL-1 transgenic mouse model and human patients using immunohistochemistry. Panel I: A representative immunohistochemistry of LN tissue from TCL-1 transgenic mouse. Lymphoid tissue were collected at wk 37 from the mouse and stained for B220 (A), CD5 (B), p-P65 (C), pSYK (D) and control antibody (E). Average of percent positive B220/CD5+ cells for pSYK and p-P65 from the transgenic mice (n = 3) is shown (F). Panel II: A representative immunohistochemistry of LN from CLL patient. LN tissue from CLL patient is stained with CD5 (A), CD19 (B), p-P65 (C), p-SYK (D) and control antibody (E). Average of percent positive CD19/CD5+ CLL cells for p-SYK and p-P65 from patient LNs (n = 3) is shown (F).
Differential Expression of Key Genes in Prognostic Groups in PB-CLL, BM-CLL and LN-CLL
We correlated gene expression profiles with three clinical parameters including chromosomal abnormalities, bulky lym-phadenopathy (BLA) and time to treatment in CLL cells from each tissue compartment to identify gene signatures that may be responsible for adverse clinical outcome.
Lymphadenopathy. GEPs derived from PB samples were compared in patients with (n = 9) or without (n = 11) BLA. Patients with BLA had a significantly higher number (n < 400) of upregulated genes (Figure 5A), and among these BCR signaling (CD79b, CD72, SYK, BTK, BLNK) was the dominant signature in this subset of cases. To confirm BCR activation, phos-phorylated-SYK (p-SYK) was measured by using flow cytometry in four samples from CLL patients with BLA and five from patients without BLA. Expression of p-SYK was significantly (p < 0.05) higher in the CLL samples with BLA compared with those without it (Figure 5B).
Chromosomal abnormalities. We performed differential expression between PB samples of patients with high-risk cytogenetic abnormalities [del(11q), del(17p) and trisomy12, n = 7] versus cases without these abnormalities (n = 12). Significantly higher expression of genes associated with BCR signaling and cell activation (BTK, AKT2, CD83, LAG3) correlated with the higher risk of genetic abnormalities (Supplementary Figure S3A). A similar observation was observed in BM-CLL GEP analysis (Supplementary Figure S3B). In addition, we observed that the transcripts of chemokine ligand CCL3 significantly correlated with higher risk of cytoge-netic abnormalities in both BM-CLL (p < 0.05) and LN-CLL (p < 0.05) specimens (Figure 5C). These observations are consistent with our validation cohort of PB-CLL samples (n = 40), where high CCL3 mRNA expression was also significantly (p = 0.02) associated with a shorter time to treatment (Figure 5D).
Time to treatment. Transcripts encoding genes associated with B-cell activation (CD69) were significantly associated with short time to treatment (<12 months; n = 9) compared with the group with a longer time to treatment (<12 months; n = 10) (Supplementary Figure S3C), consistent with the above observation. A subset of genes related to apo-ptosis (CASP6, BID, and CASP4) was underexpressed in LN cases with shorter interval of treatment, suggesting that antiapoptotic functions dominate in this group (Supplementary Figure S3D).
Overall, these observations indicate that chronic BCR signaling is a dominant pathway in CLL, as indicated by higher expression of BTK and AKT2, CD69 and phosphorylated SYK (p-SYK) in the chromosomal abnormalities, time to treatment and BLA prognostic groups, respectively, and therefore may be a valuable prognostic and therapeutic target in CLL.
Correlation of differential gene expression with a prognostic indicator such as BLA and validation of expression of key molecules such as pSyk and CCL3 expression. Gene expression profiles of PB-CLL samples were grouped on the basis of the presence and absence of BLA. (A) Gene expression signature of PB-CLL cells from BLA patients. PB-CLL samples were grouped based on a known prognostic indicator, presence or absence of BLA, and gene expression profiles of these PB-CLL samples were correlated by using significant analysis of microarray analyses. Comparison of expression profiles between PB-CLL with BLA (n = 9) and without BLA (n = 11) are shown. (B) Comparison of the expression of p-SYK in CLL samples from patients with BLA (n = 4) and those without BLA (n = 5). (C) Expression of CCL3 transcripts in BM-CLL with poor chromosomal abnormalities (ChAb) (n = 6) versus good ChAb (n = 11) and LN-CLL with poor ChAb (n = 11) versus good ChAb (n = 3). (D) Relationship between expression of CCL3 transcripts and time to treatment among the additional 40 PB-CLL samples using the Kaplan-Meier log-rank test.
Discussion
We and others have studied the role of microenvironments in survival/proliferation and migration of CLL cells using in vitro culture systems (25,29). However, in vitro culture systems have limitations and, therefore, we analyzed gene expression in CLL cells directly from these in vivo mi-croenvironments. Although the GEP of CLL cells from PB, BM and LN did not cluster in a tissue-specific manner, LN-CLL cells are far less heterogeneous than GEP of PB-CLL and BM-CLL, as evidenced by the number of clusters in Figure 2A. Our findings are consistent with the LN microenvironment providing chronic stimulation via the BCR and BAFF/APRIL, which are critical for the growth and survival of CLL cells. Our observation is consistent with the recent findings by Herishanu et al. (18) showing that the BCR signaling pathway is activated at LN sites and is likely a major pathogenetic mechanism of CLL progression (29,30,41,42). We validated this observation by measuring levels of the kinase SYK, which is a key in BCR signaling using qRT-PCR and immunohistochem-istry. Unlike Herishanu et al., our study analyzes PB, BM and LN cells from different patients, uses TCL1 transgenic mice and correlates GEP with prognostic groups. Despite using CLL cells from different sites from different patients, we observed similar findings that BCR signaling molecules are overexpressed in LN-CLL, indicating that LN tissue sites play a prominent role in influencing the gene signature. Additionally, our studies also observed that BAFF/APRIL-related genes were also enriched in LN-CLL cells. These genes have been shown to promote cell survival and adverse prognosis in CLL (31,43). These observations suggest that LN sites provide signals for chronic activation of CLL cells. Expression of p-SYK (key player of BCR signaling pathway) and p-P65 (key molecule of NF-κB pathway) in the lymphoid compartment was validated by using LN samples of patients and the Eμ-TCL1 mouse model.
Although certain interactions were specifically observed in LNs, molecules of other major pathways, for example, NF-κB and MAPK pathways (44), were constitu-tively active in all tissue sites, indicating that they may be responsible for survival/proliferation of CLL cells regardless of anatomic location. Chemokine lig-ands/receptors are being investigated extensively to better understand the migration of CLL cells. We observed that CCR7, CXCR5 and CXCR4 were overexpressed by PB-CLL, and their respective cognate ligands CCL21 and CXCL13 were expressed by LN and CXCL12 in BM sites. Upregulation of CCR7 was shown to facilitate the migration of CLL cells (32) and, similarly, it is also facilitated by the interaction of the CXCR4-CXCL12 axis in CLL (33,34). These observations indicate that distinct microenvironments express different ligands for interaction with tumor cells. Interestingly, CCL3 was expressed by all three types of CLL cells. CCL3 is known to express at high levels in CLL cells when cocultured on stroma (9,35) and does attract T cells and follicular dendritic cells, thus indicating that CCL3 is crucial for CLL cells in different microenvironments, probably to form clusters with other cell types. We observed that high expression of CCL3 was associated with the poor outcome groups in BM and LN, suggesting its role in disease progression. Consistent with these findings, higher CCL3 expression was also associated with a shorter time to treatment in PB cases. Furthermore, higher expression of CCL21 and CXCL13 was observed in the LN-CLL. These chemokine ligands are known to be expressed by CLL as well as other stromal cells, and thus a very high expression may be partially contributed because of some (<10%) contaminating stromal cells in our LN-CLL preparation.
Interestingly, an immune suppressive signature was also observed in CLL cells from LN sites as previously reported by Sasaki et al. (36) in acute-type adult T-cell leukemia. Among the key tolerogenic genes, CAV1, IFI16 and IDO1 were significantly overexpressed in LN-CLL compared with PB-CLL or BM-CLL. Although LN is an appropriate place for immune surveillance and generation of immune response, it seems to provide CLL cells with a supportive home. One explanation for this phenomenon is that the immune-sup-pressive gene signature expressed by CLL cells creates immune dysfunction in the LN site and, thus, may contribute to immune evasion of CLL cells in this site. These genes may play a crucial role in modulating T-cell behavior and immune synapse formation for the benefit of CLL cells (38,39). Because BLA is used as a marker of aggressive disease (21), we correlated GEPs with the presence and absence of BLA in PB. Interestingly, BCR signaling genes including SYK, BTK, CD79b and BLNK were overexpressed in PB-CLL from patients with BLA. Furthermore, genes related to hedgehog signaling, chemokines, BAFF/APRIL and MAPK signaling were also overexpressed in PB-CLL from BLA patients (Figure 5A). Thus, the gene expression signature of PB-CLL cells from patients with BLA resembles that of LN-CLL, suggesting that these PB-CLL cells share a close and expected relationship with the LN-CLL cells. This result is in accord with the recent reports that quiescent PB-CLL cells are similar to proliferating B cells of LN pseudofollicles (19), and a dynamic relationship exists between quiescent PB-CLL cells with the lymphoid microenvironment (37). Similarly, overexpression of BCR signaling molecules was also observed in LN and BM samples from poor prognostic groups, although the number of samples in each prognostic group was smaller in these sites.
The importance of the microenvironment has also been demonstrated by in vitro systems wherein CLL cells survive/proliferate in the presence of stro-mal cells (25). In concordance with other studies, our in vitro stromal cultures promoted survival and proliferation of CLL cells. Because the microenvironment regulates the growth of CLL cells by regulating expression of key genes (45), we studied expression of a few NF-κB-associated genes in CLL cells cocultured on stromal cells. In this study, CLL cells cultured on stroma upregulated NF-κB–associated genes IL6, BCL2 and TRAF1, thus reinforcing the importance of the microenvironment in regulating gene expression.
Conclusion
We have summarized our results and their possible interpretation in a hypothetical model of survival/proliferation and migration among PB-CLL, BM-CLL and LN-CLL (Figure 6), where it is shown that PB-CLL cells use various MAPK and NF-κB target molecules for survival/proliferation in PB and use CCR7 and CXCR5 chemokine receptors for their migration toward the LN microenvironment, mediated by their cognate ligands CCL21 and CXCL13, which are expressed by LN-CLL. In the LN, CLL cells secrete the chemokine ligand CCL3 to attract T cells and stromal cells. The interactions of CLL cells and stroma lead to the activation of BCR and BAFF/APRIL signaling, which in turn activates MAPK- and NF-κB–associated molecules. PB-CLL cells express chemokine receptor CXCR4, and its cognate ligand CXCL12/SDF-1 is secreted by BM stroma, thereby influencing the migration of PB-CLL cells to the BM.
A hypothetical model of molecular determinants of CLL cell survival, proliferation and migration in the PB, BM and LN microenvironments. Peripheral blood: CLL cells use different NF-κB- and MAPK-associated genes for survival and proliferation. PB-CLL cells express chemokine receptors, which stimulate PB-CLL migration to the LN or BM. Lymph nodes: CLL cells attract stromal cells to form a suitable microenvironment. Here, CLL cells undergo chronic activation via the BCR, BAFF/APRIL, NF-κB, MAPK and PI3K/Akt pathways, which leads to survival and proliferation. In addition, because of expression of a tolerogenic signature in LN-CLL, the immune cells are tolerogenic and unable to identify or kill CLL cells in LNs; Bone marrow: CLL cells attract stromal cells to form a suitable microenvironment. Here, CLL cells use different NF-κB and MAPK pathway genes for survival and proliferation.
In the BM, CLL cells express CCL3 to at-tract T cells and stromal cells; this interaction activates expression of NF-κB and MAPK molecules in BM-CLL cells, including v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta (NF-κBIB) and BCL2-related protein A1 (BCL2A1), for survival/proliferation. Together our results suggest that interactions between the LN microenvironment and CLL cells lead to the survival, proliferation and migration of CLL cells while inducing the host immune suppression that results in CLL progression. Overall, our study facilitates better understanding of the pathobi-ology of CLL and also lays the foundation for additional studies to eventually develop improved therapy for CLL by targeting Btk, Syk, Bcl2 and other identified key molecules. Importantly, these molecules are overexpressed in LN-CLL cells, which suggests that targeting these molecules/ LN-CLL cells residing in LN will improve better chances of disease remission.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Chiorazzi N. (2007) Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best Pract. Res. Clin. Haematol. 20:399–413.
Messmer BT, et al. (2005) In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J. Clin. Invest. 115:755–64.
Defoiche J, et al. (2008) Reduction of B cell turnover in chronic lymphocytic leukaemia. Br. J. Haematol. 143:240–7.
Chiorazzi N, Rai KR, Ferrarini M. (2005) Chronic lymphocytic leukemia. N. Engl. J. Med. 352:804–15.
Burger JA, Kipps TJ. (2002) Chemokine receptors and stromal cells in the homing and homeostasis of chronic lymphocytic leukemia B cells. Leuk. Lymphoma. 43:461–6.
Carbone A, Gloghini A, Cabras A, Elia G. (2009) The Germinal centre-derived lymphomas seen through their cellular microenvironment. Br. J. Haematol. 145:468–80.
Ayala F, Dewar R, Kieran M, Kalluri R. (2009) Contribution of bone microenvironment to leukemogenesis and leukemia progression. Leukemia. 23:2233–41.
Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. (2009) The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 114:3367–75.
Burger JA, et al. (2009) High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 113:3050–8.
Ding W, et al. (2009) Bi-directional activation between mesenchymal stem cells and CLL B-cells: implication for CLL disease progression. Br. J. Haematol. 147:471–83.
Pedersen IM, et al. (2002) Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 100:1795–801.
Tsukada N, Burger JA, Zvaifler NJ, Kipps TJ. (2002) Distinctive features of “nurselike” cells that differentiate in the context of chronic lym-phocytic leukemia. Blood. 99:1030–7.
Lapalombella R, et al. (2009) The humanized CD40 antibody SGN-40 demonstrates pre-clinical activity that is enhanced by lenalidomide in chronic lym-phocytic leukaemia. Br. J. Haematol. 144:848–55.
Mittal AK, et al. (2008) Molecular basis of proliferation/survival and migration of CLL in peripheral blood, bone marrow and lymph nodes. Blood. 112(11). ASH Annual Meeting abstract no. 546.
Herishanu Y, et al. (2008) The role of the microenvironment for CLL proliferation and survival: gene expression profiling of leukemic cells derived from blood, bone marrow and lymph nodes reveals the B-cell receptor and NF κB as dominant signaling pathways. Blood. 112(11). ASH Annual Meeting abstract no. 356.
Mittal AK, et al. (2009) Clinical heterogeneity of CLL: role for immune dysregulation mediated by the lymph node microenvironment. Blood. 114(22). ASH Annual Meeting abstract no. 1243.
Herishanu Y, et al. (2009) Comparative gene expression profiling of leukemia cells in peripheral blood and tissue compartments reveals a prominent role of the microenvironment for CLL cell proliferation. Blood. 114(22). ASH Annual Meeting abstract no. 355.
Herishanu Y, et al. (2011) The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 117:563–74.
Vandewoestyne ML, et al. (2011) Laser microdissection for the assessment of the clonal relationship between chronic lymphocytic leukemia/small lym-phocytic lymphoma and proliferating B cells within lymph node pseudofollicles. Leukemia. 25:883–8.
Joshi AD, et al. (2007) ATM, CTLA4, MNDA, and HEM1 in high versus low CD38 expressing B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 13:5295–304.
Joshi AD, et al. (2007) Bulky lymphadenopathy with poor clinical outcome is associated with ATM downregulation in B-cell chronic lympho-cytic leukemia patients irrespective of 11q23 deletion. Cancer Genet. Cytogenet. 172:120–6.
Mittal AK, et al. (2007) Molecular basis of aggressive disease in chronic lymphocytic leukemia patients with 11q deletion and trisomy 12 chromosomal abnormalities. Int. J. Mol. Med. 20:461–9.
Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. U.S.A. 98:5116–21.
Subramanian A, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102:15545–50.
Hegde GV, et al. (2008) Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: a potential new therapeutic target. Mol. Cancer. Res. 6:1928–36.
Falt S, et al. (2005) Distinctive gene expression pattern in VH3-21 utilizing B-cell chronic lym-phocytic leukemia. Blood. 106:681–9.
Hoellein A, et al. (2010) Expression of multidrug resistance-associated ABC transporters in B-CLL is independent of ZAP70 status. J. Cancer Res. Clin. Oncol. 136:403–10.
Dempsey NC, Ireland HE, Smith CM, Hoyle CF, Williams JH. (2010) Heat shock protein translocation induced by membrane fluidization increases tumor-cell sensitivity to chemotherapeutic drugs. Cancer Lett. 296:257–67.
Pleyer L, Egle A, Hartmann TN, Greil R. (2009) Molecular and cellular mechanisms of CLL: novel therapeutic approaches. Nat. Rev. Clin. Oncol. 6:405–18.
Quiroga MP, et al. (2009) B-cell antigen receptor signaling enhances chronic lymphocytic leukemia cell migration and survival: specific targeting with a novel spleen tyrosine kinase inhibitor, R406. Blood. 114:1029–37.
Bojarska-Junak A, et al. (2009) BAFF and APRIL expression in B-cell chronic lymphocytic leukemia: correlation with biological and clinical features. Leuk. Res. 33:1319–27.
Redondo-Munoz J, Jose Terol M, Garcia-Marco JA, Garcia-Pardo A. (2008) Matrix metalloproteinase-9 is up-regulated by CCL21/CCR7 interaction via extracellular signal-regulated kinase-1/2 signaling and is involved in CCL21-driven B-cell chronic lymphocytic leukemia cell invasion and migration. Blood. 111:383–6.
Burger M, et al. (2005) Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood. 106:1824–30.
Barretina J, et al. (2003) CXCR4 and SDF-1 expression in B-cell chronic lymphocytic leukemia and stage of the disease. Ann. Hematol. 82:500–5.
Sivina M, et al. (2011) CCL3 (MIP-1alpha) plasma levels and the risk for disease progression in chronic lymphocytic leukemia. Blood. 117:1662–9.
Sasaki H, et al. (2005) Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood. 105:1204–13.
Calissano C, et al. (2011) Intraclonal complexity in chronic lymphocytic leukemia: fractions enriched in recently born/divided and older/quiescent cells. Mol. Med. 17:1374–82.
Gilling CE, et al. (2012) Lymph node-induced immune tolerance in chronic lymphocytic leukaemia: a role for caveolin-1. Br. J. Haematol. 158:216–31.
Yu J, et al. (2013) Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 190:3783–97.
Efanov A, et al. (2010) CD5+CD23+ leukemic cell populations in TCL1 transgenic mice show significantly increased proliferation and Akt phos-phorylation. Leukemia. 24:970–5.
Oppezzo P, Dighiero G. (2013) Role of the B-cell receptor and the microenvironment in chronic lymphocytic leukemia. Blood Cancer J. 3:e149.
Ramsay AD, Rodriguez-Justo M. (2013) Chronic lymphocytic leukaemia: the role of the microenvironment pathogenesis and therapy. Br. J. Haematol. 162:15–24.
Cols M, et al. (2012) Stromal endothelial cells establish a bidirectional crosstalk with chronic lympho-cytic leukemia cells through the TNF-related factors BAFF, APRIL, and CD40L. J. Immunol. 188:6071–83.
Cosimo E, et al. (2013) Inhibition of NF-kappaB-mediated signaling by the cyclin-dependent ki-nase inhibitor CR8 overcomes prosurvival stimuli to induce apoptosis in chronic lymphocytic leukemia cells. Clin. Cancer Res. 19:2393–405.
Smit LA, et al. (2007) Differential Noxa/Mcl-1 balance in peripheral versus lymph node chronic lymphocytic leukemia cells correlates with survival capacity. Blood. 109:1660–8.
Acknowledgments
This work was supported by the CLL Foundation (Houston, TX, USA) and University of Nebraska Medical Center College of Medicine research funds.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it.
The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this license, visit (https://doi.org/creativecommons.org/licenses/by-nc-nd/4.0/)
About this article
Cite this article
Mittal, A.K., Chaturvedi, N.K., Rai, K.J. et al. Chronic Lymphocytic Leukemia Cells in a Lymph Node Microenvironment Depict Molecular Signature Associated with an Aggressive Disease. Mol Med 20, 290–301 (2014). https://doi.org/10.2119/molmed.2012.00303
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2012.00303