Abstract
It has long been shown that many of the presently used anticancer drugs exert their effects partly through modulating the activity of vital transcription factors. The intricacy of transcriptional regulation still represents the main obstacle for the design of transcription factor-directed agents. Systematic mapping of tumor-specific transcriptional networks and application of new molecular tools have reinforced research interest and efforts in this venue. The case of breast cancer is discussed as a representative example.
Similar content being viewed by others
Introduction
Carcinogenesis is a stepwise process whereby the accumulation of genetic and epigenetic events favors the initial formation of precancerous lesions that gradually progress to invasive carcinomas (Figure 1, upper part). A number of oncogenic transcription factors, such as activator protein (AP)-1, nuclear factor (NF)-κB and signal transducer and activator of transcription (STAT)-3/STAT-5, are overactivated in human carcinomas. Most of these transcription factors operate as double-edged swords because they contribute both to initiation and progression of carcinogenesis as well as to the development of resistance on currently applied anticancer therapies (e.g., chemotherapy, hormonal therapy, biological agents and radiotherapy). Although tumor-suppressing transcription factors, such as p53 and retinoblastoma protein (pRb), have been documented to be underactivated in carcinomas, little is known about the possibility of stimulating or stabilizing them (1). For example, progress on p53 manipulation for therapeutic purposes is limited despite the in-depth elucidation of its role in cancer formation and evolution.
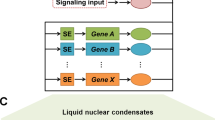
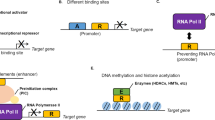
Existing versus desirable targeting of transcription factors and interconnected networks in cancer therapeutics. The uppermost schematic depicts the serial phenotypic changes and the accumulated molecular alterations (genetic and epigenetic) during carcinogenesis (breast cancer can be viewed as a typical example). This schematic has been adapted from Karamouzis et al. (40), with permission from Elsevier. BTM, basal transcriptional machinery; TF, transcription factor.
Transcription factors act through direct or indirect binding to specific DNA target sequences within gene regulatory regions. The multifaceted cross-talk between different transcription factors and their interactions with target genes across various tissues, cellular contexts and temporal settings augment the complexity of their regulatory networks. Furthermore, reversible posttranslational modifications (e.g., phosphorylations, methylations and acetylations), single-nucleotide polymorphisms and higherorder chromatin organization profoundly affect localization, turnover and genetargeting potential of transcription factors in a random-appearing fashion (2–5). Taking into consideration all of these caveats, it should be possible to either directly interfere with transcription factor DNA binding or manipulate their “wiring” within integrally cross-linked transcription modules. Up to now, transcription factor-directed anticancer drug development has focused on membrane or cytosolic targeting of molecules acting as ligand receptors. Recent technologies, such as small interfering RNA (siRNA), have shifted transcription factor targeting toward a more sophisticated, nuclear-oriented rationale (see Figure 1) (6,7).
Rewiring Transcription Factor Networks in Breast Tumors
Steroid hormone receptors are ligand-dependent intracellular transcription factors that are involved in the development and growth of several human cancers. Among the latter, breast cancer can be used as a paradigm to illustrate the complexity of transcription factor-related circuitries and the potential ways for therapeutic targeting.
Steroid hormones (e.g., estrogens) influence the development and growth of the majority of breast carcinomas (about 60% of premenopausal and 80% of post-menopausal cancers) through their binding to steroid hormone receptors (e.g., estrogen receptors [ERs]). Single-nucleotide polymorphisms in the ER gene, which may affect the binding of ER to its DNA response element and/or other cofactor proteins funneling transcription of ER target genes, have also been linked to breast carcinogenesis (8). Selective ER modulators (e.g., tamoxifen) and ER downregulators (e.g., fulvestrant) are used for the prevention and treatment of breast cancer (9). Several mechanisms of endocrine resistance have been proposed. For example, various receptor tyrosine kinases (e.g., epidermal growth factor receptor [EGFR] family members and insulin-like growth factor receptor type 1 [IGF-1R]) and nonreceptor cytoplasmic tyrosine kinases (e.g., Src kinases), as well as their downstream effectors (e.g., E2F1 transcription factor), can potentiate ER in a ligand-dependent or -independent manner (10,11). The genomic and nongenomic actions of ER are not mutually exclusive, and many crosstalk interactions have been identified. For example, resistance to hormonal therapy may be partially explained by the interplay between ER and IGF pathways. One possible mechanism by which breast cancer cells escape tamoxifen-induced apoptosis may be the activation of the AKT pathway via IGF-mediated signaling, which leads to phosphorylation of ER at serine 167 (Ser-167) and subsequent ligand-independent activation of ER (12). Additionally, IGF-binding protein 2 (IGFBP-2) mRNA and protein levels have been reported to be augmented in cell lines resistant to the anti-estrogens fulvestrant and tamoxifen (13). Selective targeting agents are being investigated in combination with endocrine therapy, in an attempt to overcome or prevent endocrine resistance in breast cancer therapeutics. Regarding IGF-1R inhibition, two different strategies have been developed: either monoclonal antibodies (mAbs) against the receptor or small molecules that impair the tyrosine kinase activity of the receptor. Another approach is the use of antisense oligonucleotides complementary to the IGF-1R mRNA region that contains the translational start site. From all the aforementioned strategies, only mAbs against the extracellular part of the receptor and inhibitors of its tyrosine kinase activity are in an advanced stage of clinical development (14,15).
AP-1 is a dimeric transcription factor comprising proteins from Jun and Fos families, for which the common denominator is the possession of basic leucine zipper (bZIP) domains that are essential for dimerization and DNA binding. Since its discovery two decades ago, AP-1 has been shown to be implicated in various aspects of tumorigenesis, including regulation of cellular proliferation and apoptosis, modulation of extracellular matrix (ECM) components as well as neoangiogenesis (see Figure 1) (16). Interfering with features of the active AP-1 transcription complexes can be exploited to attenuate breast carcinogenesis (prema-lignant lesions and in situ ductal carcinomas), in as much as this transcription factor has been demonstrated to be a crucial effector of the ERBB2 (an EGFR family member; commonly referred to as Her-2/neu) signaling cascade. Hence, a rational target is offered for the treatment of hormone receptor-negative and/or hormone-resistant breast carcinomas. Moreover, the extensive cross-talk of members of the AP-1 family (e.g., c-Fos) with other important transcription factors, such as E-twenty-six (ETS)-like protein 1 (Elk-1; a member of the ETS transcription factor family), has been documented, also revealing the role of many transcription factor coactivators (17).
The interaction between EGFR and ER signaling is complex, involving both positive and negative cross-talk, depending on the cellular context. For example, a suggested mechanism of tamoxifen resistance is triggered by the binding of tamoxifen to ER at the cell membrane and subsequent potentiation of a pathway that results in the release of membrane heparin-bound EGF and EGFR-related signaling pathway activation (18). The complicated nature of transcription factor involvement in breast carcinogenesis has been further enlarged after the identification of several microRNAs (miRNAs) that are aberrantly expressed in human breast carcinomas and participate in most of the fundamental oncogenic processes (e.g., initiation, progression, epithelial mesenchymal transition [EMT] and metastatic potential) (see Figure 1) (19,20). Each of these miRNAs hinders the expression of many genes, implying that comprehensive regulation can be achieved by antagonizing or overexpressing a single miRNA. Moreover, concomitant deregulation of these miRNAs would consequently alter the expression of a gene repertoire that either activates or inhibits each other’s transcription (transcription of any two [or more] genes within the gene repertoire), or interact directly via protein-protein interactions, thereby inducing tumorigenesis. For example, ER-positive breast tumors express higher levels of let-7 and miR-342, whereas ER-negative tumors tend to express higher levels of miR-221/222 (21). Additionally, it was demonstrated that elevated miR-206 levels contribute to EGFR-related estrogen unresponsiveness of breast cancer cells (22). Therefore, identification of miRNA targets would enable future delivery of miRNA inhibitors or miRNA mimics specifically to tumor cells. Furthermore, the combined evaluation of miRNA and gene expression may represent a way of individualizing treatment in certain breast cancer patients by predicting efficacy of individualizing treatment (23).
Taxanes are among the most commonly used chemotherapeutic agents for treating breast carcinomas. It was recently shown that the transcriptional coactivator with PDZ-binding motif oncoprotein (TAZ; also termed WWTR1), which is a major component of the Hippo-large tumor suppressor (LATS) pathway, is engaged in paclitaxel resistance (24). In addition, the expression of several genes regulated by NF-κB, such as cyclin D1, bcl-2, bcl-xl, survivin, cyclooxygenase-2 (cox-2) and X-linked inhibitor of apoptosis protein (XIAP) was reported to mediate chemo- and radioresistance (25). Moreover, the full transcriptional potentiation of NF-κB may involve the cross-talk with other signaling axes, such as EGFR, protein kinase A and phosphatidylinositol 3-kinase/AKT, and blocking the activity of these kinases can also provide an alternative strategy to fine-tune NF-κB activity in breast cancer therapeutics. Several classes of NF-κB inhibitors are currently being tested in conjunction with chemotherapy and radiotherapy. These embrace IκB kinase inhibitors, inhibitory peptides, antisense RNA and proteasome inhibitors that can impair various steps leading to NF-κB activation and sensitize tumor cells to the beneficial effects of chemotherapeutic drugs and radiation.
Gene expression profiling has categorized breast carcinomas into several subtypes that are correlated with distinct clinical outcomes and differ in treatment approach. Emerging data suggest that distinct progenitor cells in the developing mammary gland, which are characterized by the activity of specific transcription factor regulatory networks, are targeted by oncogenic events and give rise to diverse cancer subtypes. For example, the transcription factors GATA binding protein 3 (GATA-3) and fork-head box A1 (FOXA1) function with ER in mammary morphogenesis and have been found to be overexpressed in luminal type tumors (26). These transcription factors constitute a cell lineage-specific network that affects estrogen responsiveness as well as sensitivity to hormonal therapy. Despite the role of the ER-driven signaling cascade in the development of antiestrogen resistance, it is increasingly recognized that disruption of the ER/GATA-3/FOXA1 network represents an alternative way for breast cancer cells to acquire resistance to currently applied hormonal treatments (27). In addition, activation of other networks affects other cell types within the tumor microenvironment and contributes to tumor progression and metastasis (e.g., β-catenin/cAMP responsive element binding [CREB]-binding protein [CBP], chemokine receptors, retinoic acid receptors and many others) (28–32).
EMT is a normal process that is “hijacked” by breast cancer cells, which enables them to initiate systemic dissemination by downregulating epithelial cadherin expression or activity, separating cell-cell junctions, invading the surrounding tissues and intravasating the vasculature or lymphatic system (33). An array of transcription factors, including Twist1, Snail1, Snail2, zinc finger E-box-binding homeobox 1 (ZEB1), ZEB2 and members of the nuclear factor of activated T-cells (NFAT) family, have been shown to trigger EMT (34,35). Further elucidation of the molecular pathways that are involved in this process will enable targeting of specific transcription factors.
STAT-3/-5 transcription factors have been also implicated in breast cancer progression, most likely through modulating tumor microenvironment (36). Neoangiogenesis is considered a key process during tumor growth and is regulated by a balance of proangiogenic and antiangiogenic factors acting on tumor endothelial cells. Tumors express many proangiogenic factors, including vascular endothelial growth factor (VEGF), that bind to nearby dormant endothelial cells with subsequent detachment from the ECM, migration and proliferation, ultimately leading to sprouting and formation of new branches from the preexisting vasculature. Activation of the transcription factor hypoxia inducible factor (HIF) holds a central role in this process. HIF transcriptional activity is controlled by hypoxia, growth factors and related signaling pathways, cytokines and viruses (37). The Notch transcription factor family is also involved in breast carcinogenesis through modulation of tumor microenvironment and enhancement of neoangiogenesis. These effects are accomplished either by direct action or through cross-talk with other signaling pathways (e.g., ER, ERBB2) (38).
Concluding Remarks
Cancer therapeutics has been enriched in the last years with targeted biological agents against tumor cell-surface molecules and/or elements of signal transduction pathways that have been shown to be deregulated in certain carcinomas. The ever-growing understanding of the role of transcription factors and allied networks in carcinogenesis provides both a promise and a challenge for novel treatment strategies (39). Identification of the most suitable transcriptional target(s) in distinct carcinomas as well as competent nuclear-directed delivery methods are important prerequisites for the development of efficient transcription factor-targeted pharmaceuticals.
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
References
Singh S, Johnson J, Chellappan S. (2010) Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochim. Biophys. Acta. 1799:788–94.
Ozcan S, Andrali SS, Cantrell JEL. (2010) Modulation of transcription factor function by O-Glc-NAc modification. Biochim. Biophys. Acta. 1799:353–64.
Battaglia S, Maguire O, Campbell MJ. (2010) Transcription factor co-repressors in cancer biology: roles and targeting. Int. J. Cancer. 126:2511–9.
Pan Y, Tsai CJ, Ma B, Nussinov R. (2010) Mechanisms of transcription factor selectivity. Trends Genet. 26:75–83.
Huang B, Yang XD, Lamb A, Chen LF. (2010) Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 22:1282–90.
Rodriguez-Martinez JA, Peterson-Kaufman KJ, Ansari AZ. (2010) Small-molecule regulators that mimic transcription factors. Biochim. Biophys. Acta. 1799:768–74.
MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. (2011) Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 27:141–8.
Nilsson S, Gustafsson JA. (2011) Estrogen receptors: therapies targeted to receptor subtypes. Clin. Pharmacol. Ther. 89:44–55.
Ahmad N, Kumar R. (2011) Steroid hormone receptors in cancer development: a target for cancer therapeutics. Cancer Lett. 300:1–9.
Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. (2009) Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 10:709–17.
Louie MC, McClellan A, Siewit C, Kawabata L. (2010) Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol. Cancer Res. 8:343–52.
Zhang Y, et al. (2011) Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 13:R52.
Probst-Hensch NM, et al. (2010) IGFBP2 and IGFBP3 protein expressions in human breast cancer: association with hormonal factors and obesity. Clin. Cancer Res. 16:1025–32.
Carboni JM, et al. (2009) BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol. Cancer Ther. 8:3341–9.
Chakraborty AK, Welsh A, Digiovanna MP. (2010) Co-targeting the insulin-like growth factor I receptor enhances growth-inhibitory and pro-apoptotic effects of anti-estrogens in human breast cancer cell lines. Breast Cancer Res. Treat. 120:327–35.
Aguilera C, et al. (2011) c-Jun N-terminal phosphorylation antagonizes recruitment of the Mbd3/NuRD repressor complex. Nature. 469:231–5.
Zhang X, et al. (2011) Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate cFos expression in breast cancer cells. PLoS Genet. 7:1–15.
Wortham NC, et al. (2009) The DAED box protein p72 regulates ERα-/estrogen-dependent transcription and cell growth, and is associated with improved survival in ERα positive breast cancer. Oncogene. 28:4053–64.
Petrocca F, Lieberman J. (2011) Promise and challenge of RNA interference-based therapy for cancer. J. Clin. Oncol. 29:747–54.
Eades G, et al. (2011) MiR-200a regulates SIRT1 and EMT-like transformation in mammary epithelial cells. J. Biol. Chem. 286:25992–6002.
O’Day E, Lal A. (2010) MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 12:201.
Adams BD, Cowee DM, White BA. (2009) The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-α (ERα) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol. Endocrinol. 23:1215–30.
Somlo G, et al. (2011) Correlation between miRNA and gene expression profiles and response to neoadjuvant chemotherapy in patients with locally advanced and inflammatory breast cancer. J. Clin. Oncol. 29Suppl:A548.
Lai D, Ho KC, Hao Y, Yang X. (2011) Taxol resistance in breast cancer cells is mediated by the Hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 71:2728–38.
Wu Y, Zhou BP. (2010) TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br. J. Cancer. 102:639–44.
Hisamatsu Y, et al. (2011) The expression of GATA-3 and FOXA1 in breast cancer: the bio-markers of hormone sensitivity in luminal-type tumors. J. Clin. Oncol. 29Suppl:A599.
McCune K, et al. (2010) Prognosis of hormone-dependent breast cancers: implications of the presence of dysfunctional transcriptional networks activated by insulin via the immune transcription factor T-bet. Cancer Res. 70:685–96.
Ring A, Evgrafov O, Knwoles J, Kahn M. (2011) Targeting _-catenin/CBP interaction in breast cancer. J. Clin. Oncol. 29Suppl:A10516.
Kim E, et al. (2011) Biomarkers affecting metastasis and survival in paired tissues of 107 patients with metastatic breast cancer. J. Clin. Oncol. 29Suppl:A10630.
Marzese DM, et al. (2011) The relationship between the number of aberrantly methylated regions and the methylation status of p73 and RARβ genes and prognosis in patients with breast cancer. J. Clin. Oncol. 29Suppl:A10542.
Simpson N, Syed BM, Morgan DAL, Ellis IO, Cheung K. (2011) Pattern of estrogen receptor (ER)/progesterone receptor (PR)/HER2 expression in older women with primary breast cancer based on core needle biopsies and correlation with short-term clinical outcome. J. Clin. Oncol. 29Suppl:Ae21104.
Basik M, et al. (2011) Measurement of Pax2, TC21, CCND1, and RFS1 as predictive biomarkers for outcomes in the NCIC CTG MA.12 trial of tamoxifen after adjuvant chemotherapy in premenopausal women with early breast cancer. J. Clin. Oncol. 29Suppl:A560.
Giordano A, et al. (2011) Epithelial-mesenchymal transition in patients with HER2+ metastatic breast cancer. J. Clin. Oncol. 29Suppl:A623.
Siegel PM, Muller WJ. (2010) Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 29:2753–9.
Casas E, et al. (2011) Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 71:245–54.
Haftchenary S, Avadisian M, Gunning PT. (2011) Inhibiting aberrant Stat3 function with molecular therapeutics: a progress report. Anti-Cancer Drugs. 22:115–27.
Bodily JM, Mehta KP, Laimins LA. (2011) Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 71:1187–95.
Yin L, Velazquez OC, Liu ZJ. (2010) Notch signaling: emerging molecular targets for cancer therapy. Biochem. Pharmacol. 80:690–701.
Konstantinopoulos PA, Papavassiliou AG. (2011) Seeing the future of cancer-associated transcription factor drug targets. JAMA. 305:2349–50.
Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. (2007) Epigenomics in respiratory epithelium carcinogenesis: prevention and therapeutic challenges. Cancer Treat. Rev. 33:284–8.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Karamouzis, M.V., Papavassiliou, A.G. Transcription Factor Networks as Targets for Therapeutic Intervention of Cancer: The Breast Cancer Paradigm. Mol Med 17, 1133–1136 (2011). https://doi.org/10.2119/molmed.2011.00315
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2119/molmed.2011.00315