Abstract
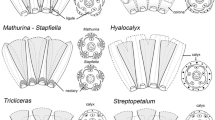
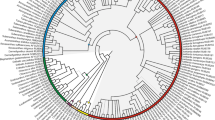
Amorpheae (Fabaceae: Papilionoideae) was first considered a natural group by Rupert Barneby in his illustrated monographDaleae Imagines. Amorpheae currently comprise eight genera, ca. 250 spp., and extensive floral diversity, including loss of corolla and addition of a stemonozone. The Amorpheae and many of Barneby’s subtribal groups are supported as monophyletic by previous phylogenetic analysis of nuclear ribosomal and chloroplast sequence data. However, some relationships remain unclear. A nuclear marker derived from a genomic study inMedicago CNGC 4, was sequenced in selected Amorpheae This is one of the first applications of this marker. for phylogenetic study. The new data confirm some relationships inferred usingtrnK and ITS, but also provide evidence for new arrangements. Combined data were used to explore several aspects of Barneby’s taxonomic framework. The phylogeny, in concert, with data on floral morphology, implies that simplification of the complex papilionoid flower has occurred several times in the history of the Amorpheae.
Similar content being viewed by others
Literature Cited
Álvarez, I. &J. F. Wendel. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417–434.
Bailey, C. D., T. G. Carr, S. A. Harris &C. E. Hughes. 2003. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Molecular Phylogenetics and Evolution 29: 435–455.
—,C. E. Hughes &S. A. Harris. 2004. Using RAPDs to identify DNA sequence loci for species level phylogeny reconstruction: an example fromLeucaena (Fabaceae). Systematic Botany 29: 4–14.
Barker, F. K. &F. M. Lutzoni. 2002. The utility of the incongruence length difference test. Systematic Biology 51: 625–637.
Barneby, R. C. 1962 A synopsis ofErrazurizia. Leaflets of Western Botany 9: 209–214.
— 1977. Daleae imagines: an illustrated revision ofErrazurizia Philippi,Psorothamnus Rydberg,Marina Liebmann, andDalea Lucanus emend. Barneby, including all species of Leguminosae tribe Amorpheae Borissova ever referred toDalea. Memoris of The New York Botanical Garden 27: 1–891.
— 1980. Three new species ofDalea sect.Parosela (Leguminosae: Amorpheae) from western and southern Mexico. Brittonia 32: 392–396.
—, 1981. New species ofDalea sectionParosela (Leguminosae: Amorpheae) from Peru and Mexico. Brittonia 33: 508–511.
— 1988. The genusDalea (Fabaceae tribe Amorpheae) in Departmento de Cajamarca, Peru, with description of three new species. Brittonia 40: 1–6.
— 1990. Two new taxa inDalea (Fabaceae: Amorpheae) from southern Mexico and northern Chile. Brittonia 42: 89–91.
Bentham, G. 1865. Leguminosae. Pages 434–600.In: G. Bentham & J. D. Hooker, editors. Genera plantarum, Lovell Reeve & Co., London, UK.
Choi, H. K., D. Kim, T. Uhm, E. Limpens, H. Lim, J.-H. Mun, P. Kalo, R. V. Penmetsa, A. Seres, O. Kulikova, B. A. Roe, T. Bisseling, G. B. Kiss &D. R. Cook. 2004a. A sequence-based genetic map ofMedicago truncatula and comparison of marker colinearity withM. sativa. Gentics 166: 1463–1502.
—. 2004b. Estimating genome conservation between crop and model legume species. Proceedings of the National Academy of Science 43: 15289–15294.
Corby, H. D. L., 1981. The systematic value of leguminous root nodules. Pages. 657–670.In: R. M. Polhili & P. H. Raven, editors. Advances in legume systematics. Part 2. Royal Botanic Gardens, Kew, UK.
Crase, D. 2004. Both: a portrait in two parts. Pantheon Books, New York, USA.
Doyle, J. J. &J. L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Driskell, A. C., C. Ané, J. G. Burleigh, M. M. McMahon, B. C. O’Meara &M. J. Sanderson. 2004. Prospects for building the tree of life from large sequence databases. Science 306: 1172–1174.
Endress, P. K., 1994. Diversity and evolutionary biology of tropical flowers. Cambridge University Press. Cambridge, UK.
Estrada-C, A. E., J. A. Villarreal-Q. &M. González-E. 2004. A new species ofDalea sect.Parosela (Fabaceae: Amorpheae) from Mexico, Brittonia 56: 67–71.
Farris, J. S., M. Källersjö, A. G. Kluge &C. Bult. 1994. Testing significance of incongruence. Cladistics 10: 315–319.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 38: 783–791.
Ferguson, I. K. 1990. The significance of some pollen morphological characters of the tribe Amorpheae and of the genusMucuna (tribe Phaseoleae) in the biology and systematics of subfamily Papilionoideae (Leguminosae). Review of Palaeobotany and Palynology 64: 129–36.
— &J. J. Skvarla. 1981. The pollen morphology of the subfamily Papilionoideae (Leguminosae). Pages 859–896.In: R. M. Polhill & P. H. Raven, editors. Advances in legume systematics. Part 2. Royal Botanic Gardens Kew, UK.
Graybeal, A., 1998. Is it better to add taxa or characters to a difficult phylogenetic problem? Systematic Biology 47: 90–17.
Guinet, P. & I. K. Ferguson. 1989. Structure, evolution, and biology of pollen in Leguminosae.In: C. H. Stirton & J. L. Zarucchi, editors. Advances in legume biology. Monographs in Systematic Botany from the Missouri Botanical Garden 29: 77–103.
Hasegawa, M., H. Kishino &T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 21: 160–174.
Hu, J.-M., M. Lavin, M. F. Wojciechowski &M. J. Sanderson. 2000. Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on chloroplasttrnK/matK sequences and its implications for evolutionary patterns in Papilionoideae. American Journal of Botany 87: 418–430.
———&—. 2002. Phylogenetic analysis of nuclear ribosomal ITS/5. 8S sequences in the tribe Millettieae (Fabaceae):Poecilanthe-Cycolobium, the core Millettieae, and theCallerya group. Systematic Botany 27: 722–733.
Hutchinson, J. 1964. The genera of flowering plants. Vol. 1. Dicotyledones Oxford University Press, Oxford, UK.
Isely, D., 1962. Leguminosae of the north-central states IV: Psoraleae. Iowa State Journal of Science 37: 103–162.
Kajita, T., H. Ohashi, Y. Tateishi, C. D. Bailey &J. J. Doyle. 2001.rbcL and legume phylogency, with particular reference to Phaseoleae, Millettieae, and allies. Systematic Botany 26: 515–536.
Lavin, M., R. T. Pennington, B. B. Klitgaard, J. I. Sprent, H. C. de Lima &P. E. Gasson. 2001. The dalbergioid legumes (Fabaceae): delimitation of a pantropical monophyletic clade. American Journal of Botnay 88: 503–533.
Maddison, W. P. 1997. Gene trees in species trees. Systematic Biology 46: 523–536.
Mansano, V. D., V. Bittrich, A. M. C. D. Tozzi &A. P. de Souza. 2004. Composition of theLecointea clade (Leguminosae, Papilionoideae, Swartzieae), a re-evaluation based on combined evidence from morphology and molecular data. Taxon 53: 1007–1018.
McMahon, M. &L. Hufford. 2002. Morphology and structural homology of corolla-androecium synorganization in the tribe Amorpheae (Fabaceae: Papilionoideae). American Journal of Botany 89: 1884–1898.
—&—. 2004. Phylogeny of Amorpheae (Fabaceae: Papilionoideae). American Journal of Botany 91: 1219–1230.
—&—. 2005. Evolution and development in the amorphoid clade (Amorpheae: Papilionoideae: Leguminosae): petal loss and dedifferentiation. International Journal of Plant Sciences 166: 383–396.
Munz, P. A., 1959. A California flora. University of California Press, Berkeley, USA.
Pennington, R. T., B. B. Klitgaard, H. Ireland &M. Lavin. 2000. New insights into floral evolution from molecular phylogenies. Pages 233–248.In: P. S. Herendeen & A. Bruneau, editors, Advances in legume systematics. Part 9. Royal Botanic Gardens, Kew, UK.
—,M. Lavin, H. Ireland, B. Klitgaard, J. Preston &J.-M. Hu. 2001. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplasttrnL intron. Systematic Botany 26: 537–556.
Polhill, R. M., 1981. Papilionoideae. Pages. 191–204.In: R. M. Polhill & P. H. Raven, editor, Advances in legume systematics. Part 1. Royal Botanic Gardens, Kew, UK.
Posada, D. &K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818.
Prenner, G. 2004. The asymmetric androecium in Papilionoideae (Leguminosae): definition, occurrence, and possible systematic value. International Journal of Plant Sciences 165: 499–510.
Rydberg, P. A., 1919. Fabaceae: Psoraleae, part 1. North American Flora 24: 1–34, 40–64.
—, 1920. Fabaceae: Psoraleae, part 2. North American Flora 24: 65–136.
— 1928a Genera of North American Fabaceae III: tribe Psoraleae. American Journal of Botany 15: 195–203.
— 1928b. Genera of North American Fabaceae IV: tribe Psoraleae (continued). American Journal of Botany 15: 425–432.
Scherson, R., R.-K. Choi, D. Cook &M. J. Sanderson. 2005. Phylogenetics of New World Astragalus: the utility of genomics technology in reconstructing phylogenies at low taxonomic levels. Britonia 57: 354–366.
Shimodaira, H. &M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116.
Shreve, F. &I. L. Wiggins. 1964. Vegetation and flora of the Sonoran Desert. Stanford University Press, Stanford, California, USA.
Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b 10. Sinauer, Sunderland, Massachusetts, USA.
Taubert, P. H. W. 1894. Galegeae Psoraliinae. Rages 263–265.In: A. Engler & K. Prantl. editors. Die Natürlichen pflanzenfamilien teile III, abteilungen 3.
Tucker, S. C. &C. H. Stirton. 1991. Development of the cymose inflorescence, cupulum and flower ofPsoralea pinnata (Leguminosae: Papilionoideae: Psoraleeae), Botanical Journal of the Linnean Society 106: 209–227.
Turner G., 1986. Comparative development of secretory cavities in the tribes Amorpheae and Psoraleeae (Leguminosae: Papilionoideae). American Journal of Botany 73: 1178–1192.
Wiggins, I. L., 1980. Flora of Baja California. Stanford University Press, Stanford, California, USA.
Wojciechowski, M. F., M. Lavin &M. J. Sanderson. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastidmatK gene resolves many well-supported subclades within the family. American Journal of Botany 91: 1846–1862.
Wolfe, A. D. &C. P. Randle. 2004. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Systematic Botany 29: 1011–1020.
Yang, Z., 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. Journal of Molecular Evolution 39: 306–314.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
McMahon, M.M. Phylogenetic relationships and floral evolution in the papilionoid legume clade Amorpheae. Brittonia 57, 397–411 (2005). https://doi.org/10.1663/0007-196X(2005)057[0397:PRAFEI]2.0.CO;2
Issue Date:
DOI: https://doi.org/10.1663/0007-196X(2005)057[0397:PRAFEI]2.0.CO;2