Abstract
Aims
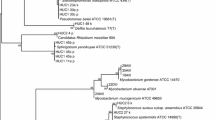
To screen for the predominant bacteria strains distributed in clean rooms and to analyze their phylogenetic relationships.
Methods and Results
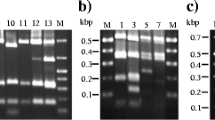
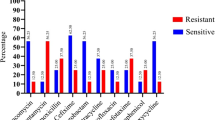
The bacteria distributed in air, surfaces and personnel in clean rooms were routinely monitored using agar plates. Five isolates frequently isolated from the clean rooms of an aseptic pharmaceutical production workshop were selected based on their colony and cell morphology characteristics. Their physiological and biochemical properties, as well as partial 16S rDNA sequences, were analyzed. Results showed that all the five isolates belong to Gram positive bacteria, of which three were Staphylococcus, one Microbacterium and one Bacillus species. Sensitivity tests for these bacteria isolates to 3 disinfectants showed that isolate F03 was obtuse, and had low susceptivity to UV irradiation, while isolates F02, F01 and F04 were not sensitive to phenol treatment. Isolates F04, F01 and F05 were resistant to chlorhexidine gluconate.
Conclusion
Bacteria widely distributed in clean rooms are mainly a group of Gram positive strains, showing high resistance to selected disinfectants. Significance and impact of the study: Clean rooms are essential in aseptic pharmaceutical and food production. Screening bacteria isolates and identifying them is part of good manufacturing practices, and will aid in finding a more effective disinfection method.
Similar content being viewed by others
References
Ajaz, M., Noor, N., Rasool, S.A., Khan, S.A., 2004. Phenol resistant-bacteria from soil: identification-characterization and genetic studies. Pakistan J. Botany, 36(2):415–424.
Akers, M.J., 1994. Parenteral Quality Control, 2nd Ed. Marcel Dekker Inc., New York.
Akers, J.E., 1997. Environmental monitoring and control: proposed standards, current practices, and future directions. J. Pharm. Sci. Technol., 51(1):36–47.
Chapman, J.S., 2003. Disinfectant resistance mechanisms, cross-resistance and co-resistance. Int. Biodeter. Biodegr., 51(4):271–276. [doi:10.1016/S0964-8305(03)00044-1]
Cloete, T.E., 2003. Resistance mechanisms of bacteria to antimicrobial compounds. Int. Biodeter. Biodegr., 51(4):277–282. [doi:10.1016/S0964-8305(03)00042-8]
Dukan, S., Touati, D., 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol., 178(21): 6145–6150.
John, G.H., Noel, R.K., Peter, H.A.S., Bergy, D., 1994. Bergey’s Manual of Determinative Bacteriology, 9th Ed. Williams & Wilkins, Lippincott.
MHPRC (Ministry of Health of People’s Republic of China), 2002. Technical Standard for Disinfection. Ministry of Health of People’s Republic of China, Beijing (in Chinese).
Nagarkar, P.P., Ravetkar, S.D., Watve, M.G., 2001. Oligophilic bacteria as tools to monitor aseptic pharmaceutical production units. Appl. Environ. Microbiol., 67(3): 1371–1374. [doi:10.1128/AEM.67.3.1371-1374.2001]
Newcombe, D.A., Schuerger, A.C., Benardini, J.N., Dickinson, D., Tanner, R., Venkateswaran, K., 2005. Survival of spacecraft-associated microorganisms under simulated martian UV irradiation. Appl. Environ. Microbiol., 71(12): 8147–8156. [doi:10.1128/AEM.71.12.8147-8156.2005]
Ogunniyi, T.A.B., Oni, P.O., Juba, A., Asaolu, S.O., Kolawole, D.O., 2000. Disinfectants/antiseptics in the management of guinea worm ulcers in the rural areas. Acta Tropica, 74(1):33–38. [doi:10.1016/S0001-706X(99)00057-1]
Onaolapo, J.A., 1995. Contamination of hospital disinfectants and antiseptics. Int. Biodeter. Biodegr., 36(3-4):471–472. [doi:10.1016/0964-8305(96)81852-X]
Park, H.D., Noguera, D.R., 2007. Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J. Appl. Microbiol., 102(5):1401–1417. [doi:10.1111/j.1365-2672.2006.03176.x]
Rosch, P., Harz, M., Schmitt, M., Peschke, K.D., Ronneberger, O., Burkhardt, H., Motzkus, H.W., Lankers, M., Hofer, S., Thiele, H., Popp, J., 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol., 71(3):1626–1637. [doi:10.1128/AEM.71.3.1626-1637.2005]
Schicht, H.H., 1998. New European GMP rules for manufacturing sterile medicinal products. Cleanrooms Int., 2:17–28.
Shaban, A.M., El-Taweel, G.E., Ali, G.H., 1997. UV ability to inactivate microorganisms combined with factors affecting radiation. Wat. Sci. Technol., 35(11-12):107–112. [doi:10.1016/S0273-1223(97)00243-6]
Teske, A., Sigalevich, P., Cohen, Y., Muyzer, G., 1996. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl. Environ. Microbiol., 62(11):4210–4215.
USGSA (US General Services Administration), 1992. Federal Standards 209E: Airborne Particulate Cleanliness Classes in Clean Rooms and Clean Zones. General Services Administration, Washington DC.
Wu, G.F., Zhou, X.P., 2005. Characterization of phosphorus-releasing bacteria in a small eutrophic shallow lake, Eastern China. Wat. Res., 39(19):4623–4632. [doi:10.1016/j.watres.2005.08.036]
Author information
Authors and Affiliations
Additional information
Project (No. 30371703) supported by the National Natural Science Foundation of China
Rights and permissions
About this article
Cite this article
Wu, Gf., Liu, Xh. Characterization of predominant bacteria isolates from clean rooms in a pharmaceutical production unit. J. Zhejiang Univ. - Sci. B 8, 666–672 (2007). https://doi.org/10.1631/jzus.2007.B0666
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1631/jzus.2007.B0666