Abstract
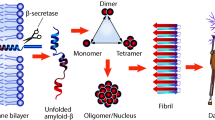
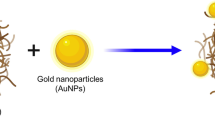
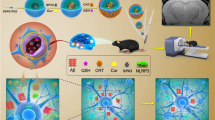
Numerous studies have reported that amyloid-beta 42 (Aβ-42) protein is a high-profile risk factor associated with the onset and progression of Alzheimer’s disease (AD). Accumulation of extracellular senile plaques, synaptic degeneration, and intracellular neurofibrillary tangles were recorded as essential features that facilitate the onset of Aβ-42, resulting in AD. Hence, we attempted a new screening technique to discover potential inhibitors against Aβ-42 using an in silico deep neural network approach. We screened PubChem compounds library and found wgx-50 as a potential inhibitor of Aβ-42. Also, synergistic effects of wgx-50–gold nanoparticles (AuNPs) complex induced significant inhibition of Aβ-42, compared with those of wgx-50 alone. Further, molecular docking analysis, systems biology approach, and time course simulation confirmed that synergistic effects of wgx-50–AuNPs complex have potential application in the treatment for AD. Additionally, we proposed the biological circuit for AD induced by Aβ-42 that can be used to monitor the effect of drugs on AD.





Similar content being viewed by others
References
A.C. Kaushik, A. Kumar, V.D. Dwivedi, S. Bharadwaj, S. Kumar, K. Bharti, P. Kumar, R.K. Chaudhary, S.K. Mishra: Deciphering the biochemical pathway and pharmacokinetic study of amyloid β-42 with superparamagnetic iron oxide nanoparticles (SPIONs) using systems biology approach. Mol. Neurobiol. 55, 3224–3236 (2018).
B.M. Austen, E.R. Frears, and H. Davies: The use of Seldi ProteinChip™ arrays to monitor production of Alzheimer’s β-amyloid in transfected cells. J. Pept. Sci. 6, 459–469 (2000).
D. Beher, J.D. Wrigley, A.P. Owens, and M.S. Shearman: Generation of C-terminally truncated amyloid-β peptides is dependent on γ-secretase activity. J. Neurochem. 82, 563–575 (2002).
J. Marksteiner, H. Hinterhuber, and C. Humpel: Cerebrospinal fluid biomarkers for diagnosis of Alzheimer’s disease: Beta-amyloid (1-42), tau, phospho-tau-181 and total protein. Drugs Today 43, 423 (2007).
R. Brookmeyer, E. Johnson, K. Ziegler-Graham, H.M. Arrighi: Forecasting the global burden of Alzheimer’s disease. Alzheimers Dement. 3, 186–191 (2007).
M. Goedert and M.G. Spillantini: A century of Alzheimer’s disease. Science 314, 777–781 (2006).
T.C. Saido, T. Iwatsubo, D.M. Mann, H. Shimada, Y. Ihara, S. Kawashima: Dominant and differential deposition of distinct β-amyloid peptide species, AβN3(pE), in senile plaques. Neuron 14, 457–466 (1995).
J.A. Hardy and G.A. Higgins: Alzheimer’s disease: The amyloid cascade hypothesis. Science 256, 184–185 (1992).
R. Kayed, E. Head, J.L. Thompson, T.M. McIntire, S.C. Milton, C.W. Cotman, C.G. Glabe: Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
M. Kawahara and Y. Kuroda: Molecular mechanism of neurodegeneration induced by Alzheimer’s beta-amyloid protein: Channel formation and disruption of calcium homeostasis. Brain Res. Bull. 53, 389–397 (2000).
A. Takashima, K. Noguchi, K. Sato, T. Hoshino, K. Imahori: Tau protein kinase I is essential for amyloid beta-protein-induced neurotoxicity. Proc. Natl. Acad. Sci. U. S. A. 90, 7789–7793 (1993).
S.M. Yatin, M. Aksenova, M. Aksenov, W.R. Markesbery, T. Aulick, and D.A. Butterfield: Temporal relations among amyloid beta-peptide-induced free-radical oxidative stress, neuronal toxicity, and neuronal defensive responses. J. Mol. Neurosci. 11, 183–197 (1998).
H.W. Querfurth, J. Jiang, J.D. Geiger, D.J. Selkoe: Caffeine stimulates amyloid beta-peptide release from beta-amyloid precursor protein-transfected HEK293 cells. J. Neurochem. 69, 1580–1591 (1997).
R. Capone, H. Jang, S.A. Kotler, L. Connelly, F. Teran Arce, S. Ramachandran, B.L. Kagan, R. Nussinov, R. Lal: All-D-enantiomer of beta-amyloid peptide forms ion channels in lipid bilayers. J. Chem. Theory Comput. 8, 1143–1152 (2012).
A. Quist, I. Doudevski, H. Lin, R. Azimova, D. Ng, B. Frangione, B. Kagan, J. Ghiso, R. Lal: Amyloid ion channels: A common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. U. S. A. 102, 10427–10432 (2005).
R. Bhatia, H. Lin, and R. Lal: Fresh and globular amyloid β protein (1-42) induces rapid cellular degeneration: Evidence for AβP channel-mediated cellular toxicity. FASEB J. 14, 1233–1243 (2000).
M. Lu, M.K. Jolly, and E. Ben-Jacob: Toward decoding the principles of cancer metastasis circuits. Cancer Res. 74, 4574–4587 (2014).
M.K. Jolly, B. Huang, M. Lu, S.A. Mani, H. Levine, and E. Ben-Jacob: Towards elucidating the connection between epithelial–mesenchymal transitions and stemness. J. R. Soc. Interface 11, 20140962 (2014).
C. Kiel, E. Yus, and L. Serrano: Engineering signal transduction pathways. Cell 140, 33–47 (2010).
D. McMillen, N. Kopell, J. Hasty, and J. Collins: Synchronizing genetic relaxation oscillators by intercell signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 679–684 (2002).
D. Densmore and S. Hassoun: Design automation for synthetic biological systems. IEEE Des. Test Comput. 29, 7–20 (2012).
A.S. Khalil and J.J. Collins: Synthetic biology: Applications come of age. Nat. Rev. Genet. 11, 367–379 (2010).
E.M. Ozbudak, M. Thattai, I. Kurtser, A.D. Grossman, and A. Van Oudenaarden: Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73 (2002).
M. Tang, Z. Wang, Y. Zhou, W. Xu, S. Li, L. Wang, D. Wei, Z. Qiao: A novel drug candidate for Alzheimer’s disease treatment: wgx-50 derived from zanthoxylum bungeanum. J. Alzheim. Dis. 34, 203–213 (2013).
S. Hou, R.X. Gu, and D.Q. Wei: Inhibition of beta-amyloid channels with a drug candidate wgx-50 revealed by molecular dynamics simulations. J. Chem. Inf. Model. 57, 2811–2821 (2017).
H.M. Fan, R.X. Gu, Y.J Wang, Y.L. Pi, Y.H. Zhang, Q. Xu, D.Q. Wei: Destabilization of Alzheimer’s Aβ42 protofibrils with a novel drug candidate wgx-50 by molecular dynamics simulations. J. Phys. Chem. B. 119, 11196–11202 (2015).
A.C. Kaushik, S. Sahi: Boolean network model for GPR142 against Type 2 diabetes and relative dynamic change ratio analysis using systems and biological circuits approach. Systems and synthetic biology 1; 9(1-2): 45–54 (2015).
L.M. Adleman: Molecular computation of solutions to combinatorial problems. Nature 369, 40 (1994).
D. Boneh, C. Dunworth, R.J. Lipton, and J. Sgall: On the computational power of DNA. Discrete Appl. Math. 71, 79–94 (1996).
L. Kari, G. Gloor, and S. Yu: Using DNA to solve the bounded post correspondence problem. Theor. Comput. Sci. 231, 193–203 (2000).
M. Ogihara and A. Ray: Simulating Boolean circuits on a DNA computer. Algorithmica 25, 239–250 (1999).
Y. Benenson, B. Gil, U. Ben-Dor, R. Adar, and E. Shapiro: An autonomous molecular computer for logical control of gene expression. Nature 429, 423–429 (2004).
PubChem Home Page: Available at: http://pubchem.ncbi.nlm.nih.gov/ (accessed November 22, 2017).
G.M. Morris, R. Huey, W. Lindstrom, M.F. Sanner, R.K. Belew, D.S. Goodsell, and A.J. Olson: AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
A.C. Kaushik, S. Bharadwaj, S. Kumar, D.Q. Wei: Nano-particle mediated inhibition of Parkinson’s disease using computational biology approach. Sci. Rep. 8, 9169 (2018).
C. Burch: Logisim a graphical system for logic circuit design and simulation. J. Educ. Resour. Comput. 2, 5–16 (2002).
Acknowledgments
The simulations in this work were supported by the Center for High Performance Computing, Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Corresponding author
Supplementary material
Rights and permissions
About this article
Cite this article
Kaushik, A.C., Kumar, A., Peng, Z. et al. Evaluation and validation of synergistic effects of amyloid-beta inhibitor–gold nanoparticles complex on Alzheimer’s disease using deep neural network approach. Journal of Materials Research 34, 1845–1853 (2019). https://doi.org/10.1557/jmr.2018.452
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1557/jmr.2018.452