Abstract
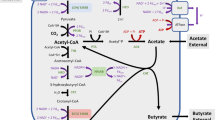
Clostridium thermosuccinogenes are anaerobic thermophilic bacteria that ferment various carbohydrates to succinate and acetate as major products and formate, lactate, and ethanol as minor products. Metabolic carbon flux analysis was used to evaluate the effect of pH and redox potential on the batch fermentation of C. thermosuccinogenes. In a first study, the effects of four pH values (6.50, 6.75, 7.00, and 7.25) on intracellular carbon flux at a constant redox potential of −275 mV were compared. The flux of carbon toward succinate and formate increased whereas the flux to lactate decreased significantly with a pH increase from 6.50 to 7.25. Both specific growth rate and specific rate of glucose consumption were unaffected by changes in pH. The fraction of carbon flux at the phosphoenolpyruvate (PEP) node flowing to oxaloacetate increased with an increase in pH. At the pyruvate node, the fraction of flux to formate increased with increasing pH. At the acetyl CoA node, the fraction of flux to acetate increased significantly with an increase in pH. A second study elucidated the effect of four controlled culture redox potentials (−225, −250, −275, and −310 mV) on metabolic carbon flux at a constant pH of 7.25. Lower values of culture redox potential were correlated with increased succinate, acetate, and formate fluxes and decreased ethanol and hydrogen fluxes in C. thermosuccinogenes. Lactate formation was not significantly influenced by redox potential. At the PEP node, the fraction of carbon to oxaloacetate increased with a decrease in redox potential. At the pyruvate node, the fraction of carbon to formate increased, while at the acetyl CoA node, the fraction of carbon flux to acetate increased with reduced redox potential. The presence of hydrogen in the headspace or the addition of nicotinic acid to the growth media resulted in increased hydrogen and ethanol fluxes and decreased succinate, acetate, formate, and lactate fluxes.
Similar content being viewed by others
References
Datta, R., Glassner, D. A., Jain, M. K., and Vick Roy, J. R. (1991), European patent 405,707.
Gokarn, R. R., Eiteman, M. A., and Sridhar, J. (1997), ACS Symp. Ser. 666, 237–253.
Zeikus, J. G., Elankovan, P., and Grethlein, A. (1995), Chem. Proc. 58, 71–73.
Datta, R. (1989), US patent 4,885,247.
Glassner, D. A. (1989), European patent 389,103.
Glassner, D. A. and Datta, R. (1992), US patent 5,143,834.
Guettler, M. V., Jain, M. K., and Soni, B. K. (1996), US patent 5,504,004.
Drent, W. J., Lahpor, G. A., Wiegant, W. M., and Gottschal, J. C. (1991), Appl. Environ. Microbiol. 57, 455–462.
Montville, T. J., Parris, N., and Conway, L. K. (1985), Appl. Environ. Microbiol. 49, 733–736.
Samuelov, N. S., Lamed, R., Lowe, S., and Zeikus, J. G. (1991), Appl. Environ. Microbiol. 57, 3013–3019.
Shibai, H., Ishizak, A., Kobayshi, K., and Hirose, Y. (1974), Agric. Biol. Chem. 38, 2407–2411.
Jee, H. S., Mano, T., Nishio, N., and Nagai, S. (1987), J. Gen. Appl. Microbiol. 33, 401–408.
Jee, H. S., Mano, T., Nishio, N., and Nagai, S. (1988), J. Ferment. Technol. 66, 123–126.
Kim, T. S. and Kim, B. H. (1988), Biotechnol. Lett. 10, 123–128.
Aiba, S. and Matsuoka, M. (1979), Biotechnol. Bioeng. 21, 1373–1386.
Chao, P.-Y., and Liao, J. C. (1993), Appl. Environ. Microbiol. 59, 4261–4265.
Diaz-Ricci, J. C., Tsu, M., and Bailey, J. E. (1992), Biotechnol. Bioeng. 38, 1318–1324.
Goel, A., Ferrance, J., Jeong, J., and Ataai, M. M. (1993), Biotechnol. Bioeng. 42, 686–696.
Vallino, J. J. and Stephanopoulos, G. (1993), Biotechnol. Bioeng. 41, 633–646.
Reardon, K. F., Scheper, T., and Bailey, J. E. (1987), Biotechnol. Prog. 3, 153–167.
Abbad-Andaloussi, S., Durr, C., Raval, G., and Petitdemange, H. (1996), Microbiology 142, 1149–1158.
Venkatesh, K. V. (1997), Proc. Biochem. 32, 651–655.
Sridhar, J., Eiteman, M. A., and Wiegel, J. W. (2000), Appl. Environ. Microbiol. 66, 246–251.
Sridhar, J. and Eiteman, M. A. (1999), Appl. Biochem. Biotechnol. 82, 91–101.
Eiteman, M. A. and Chastain, M. J. (1997), Anal. Chim. Acta 338, 69–75.
Ott, L. (1993), An Introduction to Statistical Methods and Data Analysis, 4th ed., Wadsworth, Belmont, CA.
Gottschalk, G. (1986), in Bacterial Metabolism, Springer-Verlag, New York, pp. 210–280.
Erickson, L. E. (1980), Biotechnol. Bioeng. 22, 451–456.
Park, S. M., Sinskey, A. J., and Stephanopoulos, G. (1997), Biotechnol. Bioeng. 55, 864–879.
Cook, G. M., Russell, J. B., Reichert, A., and Wiegel, J. (1996), Appl. Environ. Microbiol. 62, 4576–4579.
Cook, G. M., Janssen, P. H., and Morgan, H. W. (1993), Appl. Environ. Microbiol. 59, 2984–2990.
Stephanopoulos, G. N., Aristidou, A. A., and Nielson, J. (1998), Metabolic Engineering: Principles and Methodologies, Academic, New York.
Niedhardt, F. C., Ingraham, J. L., and Schaechter, M. (1990), Physiology of the Bacterial Cell: A Molecular Approach, Sinauer Associates, Sunderland, MA.
Tsai, S. P. and Lee, Y. H. (1988), Biotechnol. Bioeng. 32, 713–715.
Peguin, S. and Soucaille, P. (1996), Biotechnol. Bioeng. 51, 342–348.
Guedon, E., Payot, S., Desvaux, M., and Petitdemange, H. (1999), J. Bacteriol. 181, 3262–3269.
Alam, K. Y. and Clark, D. P. (1989), J. Bacteriol. 171, 6213–6217.
Leonardo, M. R., Dailly, Y., and Clark, D. P. (1996), J. Bacteriol. 178, 6013–6020.
Lovitt, R. W., Shen, G.-J., and Zeikus, J. G. (1988), J. Bacteriol. 170, 2809–2815.
Snoep, J. L., De Graef, M. R., Joost Teixeria De Mattos, M., and Neijssel, O. M. (1992), J. Gen. Microbiol. 138, 2015–2020.
London, J. and Knight, M. (1966), J. Gen. Microbiol. 44, 241–254.
Riebling, V., Thauer, R. K., and Jungermann, K. (1975), Eur. J. Biochem. 55, 445–453.
Huang, L., Forsberg, C. W., and Gibbins, L. N. (1986), Appl. Environ. Microbiol. 51, 1230–1234.
Utter, M. F. and Kolenbrander, H. M. (1972), in The Enzymes, vol. 6, 3rd ed., Boyer, P. D., ed., Academic, New York, pp. 117–165.
Jones, R. P. and Greenfield, P. F. (1982), Enzyme Microbiol. Technol. 4, 210–223.
Takai, K., Sako, Y., Uchida, A., and Ishida, Y. (1997), J. Biochem. 122, 32–40.
Turenen, M., Parkinnen, E., Londesborough, J., and Korhola, M. (1987), J. Gen. Microbiol. 133, 2865–2873.
Blackwood, A. C., Neish, A. C., and Ledingham, G. A. (1957), J. Bacteriol. 72, 497–499.
Snoep, J. L., Joost Teixeira de Mattos, M., Postma, P. W., and Niejssel, O. M. (1990), Arch. Microbiol. 154, 50–55.
Thauer, R. K., Kichniawy, F. H., and Jungermann, K. A. (1972), Eur. J. Biochem. 27, 282–290.
Klotzsch, H. R. (1969), Methods Enzymol. 13, 381–386.
Diez-Gonzalez, F., Russell, J. B., and Hunter, J. B. (1997), Arch. Microbiol. 166, 418–420.
Yan, R. and Chen, J. S. (1990), Appl. Environ. Microbiol. 56, 2591–2599.
Chen, J.-S. (1995), FEMS Microbiol. Rev. 17, 263–273.
Millay, R. H. and Hersh, L. B. (1976), J. Biol. Chem. 251, 2754–2760.
Clark, D. P. (1989), FEMS Microbiol. Rev. 63, 223–234.
Lindmark, D. G., Paolella, P., and Wood, N. P. (1969), J. Biol. Chem. 13, 3605–3612.
Vasconcelos, I., Girbal, L., and Soucaille, P. (1994), J. Bacteriol. 176, 1443–1450.
Baut, F., Fick, M., Viriot, M. L., Andre, J. C., and Engasser, J. M. (1994), Appl. Microbiol. Biotechnol. 41, 551–555.
Garrigues, C., Loubiere, P., Lindley, N. D., and Cocaign-Bousquet, M. (1997), J. Bacteriol. 179, 5282–5287.
Girbal, L. and Soucaille, P. (1994), J. Bacteriol. 176, 6433–6438.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sridhar, J., Eiteman, M.A. Metabolic flux analysis of clostridium thermosuccinogenes . Appl Biochem Biotechnol 94, 51–69 (2001). https://doi.org/10.1385/ABAB:94:1:51
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1385/ABAB:94:1:51