Abstract
Background
Current risk assessment tools to estimate the risk of nonsentinel lymph node metastases after completion lymphadenectomy for a positive sentinel lymph node (SLN) biopsy in cutaneous melanoma are based on clinical and pathologic factors. We identified a novel genetic signature that can predict non-SLN metastases in patients with cutaneous melanoma staged with a SLN biopsy.
Methods
RNA was collected for tumor-positive SLNs in patients staged by SLN biopsy for cutaneous melanoma. All patients with a tumor-positive SLN biopsy underwent completion lymphadenectomy. A 1:10 case:control series of positive and negative non-SLN patients was analyzed by microarray and quantitative RT-PCR. Candidate differentially expressed genes were validated in a 1:3 case:control separate cohort of positive and negative non-SLN patients.
Results
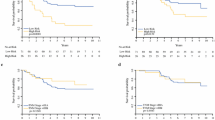
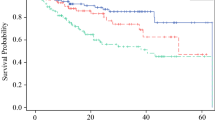
The 1:10 case:control discovery set consisted of 7 positive non-SLN cases matched to 70 negative non-SLN controls. The cases and controls were similar with regards to important clinicopathologic factors, such as gender, primary tumor site, age, ulceration, and thickness. Microarray and RT-PCR identified six potential differentially expressed genes for validation. In the 40-patient separate validation set, 10 positive non-SLN patients were matched to 30 negative non-SLN controls based on gender, ulceration, age, and thickness. Five of the six genes were differentially expressed. The five gene panel identified patients at low (7.1%) and high risk (66.7%) for non-SLN metastases.
Conclusions
A novel, non-SLN gene score based on differential expressed genes in a tumor-positive SLN can identify patients at high and low risk for non-SLN metastases.


Similar content being viewed by others
References
Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–206.
Wrightson WR, Wong SL, Edwards MJ, et al. Complications associated with sentinel lymph node biopsy for melanoma. Ann Surg Oncol. 2003;10(6):676–80.
Morton DL, Cochran AJ, Thompson JF, et al. Sentinel node biopsy for early-stage melanoma: accuracy and morbidity in MSLT-I, an international multicenter trial. Ann Surg. 2005;242(3):302–11.
Egger ME, Callender GG, McMasters KM, et al. Diversity of stage III melanoma in the era of sentinel lymph node biopsy. Ann Surg Oncol. 2013;20(3):956–63.
Rossi CR, De Salvo GL, Bonandini E, et al. Factors predictive of nonsentinel lymph node involvement and clinical outcome in melanoma patients with metastatic sentinel lymph node. Ann Surg Oncol. 2008;15(4):1202–10.
Leiter U, Stadler R, Mauch C, et al. Complete lymph node dissection versus no dissection in patients with sentinel lymph node biopsy positive melanoma (DeCOG-SLT): a multicentre, randomised, phase 3 trial. Lancet Oncol. 2016;17(6):757–67.
Faries MB, Thompson JF, Cochran AJ, et al. Completion dissection or observation for sentinel-node metastasis in melanoma. N Engl J Med. 2017;376(23):2211–22.
Leung AM, Morton DL, Ozao-Choy J, et al. Staging of regional lymph nodes in melanoma: a case for including nonsentinel lymph node positivity in the American Joint Committee on Cancer staging system. JAMA Surg. 2013;148(9):879–84.
Reintgen M, Murray L, Akman K, et al. Evidence for a better nodal staging system for melanoma: the clinical relevance of metastatic disease confined to the sentinel lymph nodes. Ann Surg Oncol. 2013;20(2):668–74.
Brown RE, Ross MI, Edwards MJ, et al. The prognostic significance of nonsentinel lymph node metastasis in melanoma. Ann Surg Oncol. 2010;17(12):3330–5.
Weber J, Mandala M, Del Vecchio M, et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N Engl J Med. 2017;377(19):1824–35.
Long GV, Hauschild A, Santinami M, et al. adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med. 2017;377(19):1813–23.
Eggermont AM, Chiarion-Sileni V, Grob JJ, et al. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N Engl J Med. 2016;375(19):1845–55.
Murali R, Desilva C, Thompson JF, Scolyer RA. Non-sentinel node risk score (N-SNORE): a scoring system for accurately stratifying risk of non-sentinel node positivity in patients with cutaneous melanoma with positive sentinel lymph nodes. J Clin Oncol. 2010;28(29):4441–9.
Feldmann R, Fink AM, Jurecka W, Rappersberger K, Steiner A. Accuracy of the non-sentinel node risk score (N-SNORE) in patients with cutaneous melanoma and positive sentinel lymph nodes: a retrospective study. Eur J Surg Oncol. 2013. https://doi.org/10.1016/j.ejso.2013.08.022.
Roka F, Mastan P, Binder M, et al. Prediction of non-sentinel node status and outcome in sentinel node-positive melanoma patients. Eur J Surg Oncol. 2008;34(1):82–8.
Gershenwald JE, Andtbacka RH, Prieto VG, et al. Microscopic tumor burden in sentinel lymph nodes predicts synchronous nonsentinel lymph node involvement in patients with melanoma. J Clin Oncol. 2008;26(26):4296–303.
van der Ploeg AP, van Akkooi AC, Haydu LE, et al. The prognostic significance of sentinel node tumour burden in melanoma patients: An international, multicenter study of 1539 sentinel node-positive melanoma patients. Eur J Cancer. 2014;50(1):111–20.
van Akkooi AC, Nowecki ZI, Voit C, et al. Sentinel node tumor burden according to the Rotterdam criteria is the most important prognostic factor for survival in melanoma patients: a multicenter study in 388 patients with positive sentinel nodes. Ann Surg. 2008;248(6):949–55.
van der Ploeg AP, van Akkooi AC, Rutkowski P, et al. Prognosis in patients with sentinel node-positive melanoma is accurately defined by the combined Rotterdam tumor load and Dewar topography criteria. J Clin Oncol. 2011;29(16):2206–14.
Cadili A, Dabbs K, Scolyer RA, Brown PT, Thompson JF. Re-evaluation of a scoring system to predict nonsentinel-node metastasis and prognosis in melanoma patients. J Am Coll Surg. 2010;211(4):522–5.
Egger ME, Bower MR, Czyszczon IA, et al. Comparison of sentinel lymph node micrometastatic tumor burden measurements in melanoma. J Am Coll Surg. 2014;218(4):519–28.
McMasters KM, Noyes RD, Reintgen DS, et al. Lessons learned from the Sunbelt Melanoma Trial. J Surg Oncol. 2004;86(4):212–23.
Hao H, Xiao D, Pan J, et al. Sentinel lymph node genes to predict prognosis in node-positive melanoma patients. Ann Surg Oncol. 2017;24(1):108–16.
Fleiss JL, Levin B, Paik MC. Statistical methods for rates and proportions statistical methods for rates and proportions, 3rd edn. Hoboken: Wiley, 2003.
Agresti A. Categorical data analysis, 2nd edn. New York: Wiley; 2002.
Gonen M. Receiver operator characteristics (ROC) curves. Proceedings of the 31st Annual SAS Users Group International Conference. 2006;120:210–31.
McMasters KM, Wong SL, Edwards MJ, et al. Frequency of nonsentinel lymph node metastasis in melanoma. Ann Surg Oncol. 2002;9(2):137–41.
Dewar DJ, Newell B, Green MA, Topping AP, Powell BW, Cook MG. The microanatomic location of metastatic melanoma in sentinel lymph nodes predicts nonsentinel lymph node involvement. J Clin Oncol. 2004;22(16):3345–49.
Scolyer RA, Li LX, McCarthy SW, et al. Micromorphometric features of positive sentinel lymph nodes predict involvement of nonsentinel nodes in patients with melanoma. Am J Clin Pathol. 2004;122(4):532–9.
Fink AM, Weihsengruber F, Duschek N, et al. Value of micromorphometric criteria of sentinel lymph node metastases in predicting further nonsentinel lymph node metastases in patients with melanoma. Melanoma Res. 2011;21(2):139–43.
Scoggins CR, Ross MI, Reintgen DS, et al. Prospective multi-institutional study of reverse transcriptase polymerase chain reaction for molecular staging of melanoma. J Clin Oncol. 2006;24(18):2849–57.
Hilari JM, Mangas C, Xi L, et al. Molecular staging of pathologically negative sentinel lymph nodes from melanoma patients using multimarker, quantitative real-time rt-PCR. Ann Surg Oncol. 2009;16(1):177–85.
Gonzalez Cao M, Badenas C, Malvehy J, et al. Prognostic value of tyrosinase reverse transcriptase PCR analysis in melanoma sentinel lymph nodes: long-term follow-up analysis. Clin Exp Dermatol. 2009;34(8):863–9.
Torisu-Itakura H, Lee JH, Scheri RP, et al. Molecular characterization of inflammatory genes in sentinel and nonsentinel nodes in melanoma. Clin Cancer Res. 2007;13(11):3125–32.
Vallacchi V, Vergani E, Camisaschi C, et al. Transcriptional profiling of melanoma sentinel nodes identify patients with poor outcome and reveal an association of CD30(+) T lymphocytes with progression. Cancer Res. 2014;74(1):130–40.
Camisaschi C, Vallacchi V, Castelli C, Rivoltini L, Rodolfo M. Immune cells in the melanoma microenvironment hold information for prediction of the risk of recurrence and response to treatment. Expert Rev Mol Diagn. 2014;14(6):643–6.
Mohos A, Sebestyen T, Liszkay G, et al. Immune cell profile of sentinel lymph nodes in patients with malignant melanoma-FOXP3 + cell density in cases with positive sentinel node status is associated with unfavorable clinical outcome. J Transl Med. 2013;11:43.
Xing Y, Nakamura Y, Rainey WE. G protein-coupled receptor expression in the adult and fetal adrenal glands. Mol Cell Endocrinol. 2009;300(1-2):43–50.
Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–73.
Cui X, Song B, Hou L, Wei Z, Tang J. High expression of osteoglycin decreases the metastatic capability of mouse hepatocarcinoma Hca-F cells to lymph nodes. Acta Biochim Biophys Sin (Shanghai). 2008;40(4):349–55.
Cheung IY, Feng Y, Danis K, et al. Novel markers of subclinical disease for Ewing family tumors from gene expression profiling. Clin Cancer Res. 2007;13(23):6978–83.
Xu L, Shen SS, Hoshida Y, et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6(5):760–9.
Mithani SK, Smith IM, Califano JA. Use of integrative epigenetic and cytogenetic analyses to identify novel tumor-suppressor genes in malignant melanoma. Melanoma Res. 2011;21(4):298–307.
Burnett RM, Craven KE, Krishnamurthy P, et al. Organ-specific adaptive signaling pathway activation in metastatic breast cancer cells. Oncotarget. 2015;6(14):12682–96.
Yang R, Chen B, Pfutze K, et al. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis. 2014;35(2):315–23.
Fimia GM, De Cesare D, Sassone-Corsi P. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol Cell Biol. 2000;20(22):8613–22.
Roger P, Pujol P, Lucas A, Baldet P, Rochefort H. Increased immunostaining of fibulin-1, an estrogen-regulated protein in the stroma of human ovarian epithelial tumors. Am J Pathol. 1998;153(5):1579–88.
Greene LM, Twal WO, Duffy MJ, et al. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br J Cancer. 2003;88(6):871–8.
Cooley MA, Kern CB, Fresco VM, et al. Fibulin-1 is required for morphogenesis of neural crest-derived structures. Dev Biol. 2008;319(2):336–45.
Cui Y, Liu J, Yin HB, Liu YF, Liu JH. Fibulin-1 functions as a prognostic factor in lung adenocarcinoma. Jpn J Clin Oncol. 2015;45(9):854–9.
Zhu J, Chen R, Mo L, et al. Expression of fibulin-1 predicted good prognosis in patients with colorectal cancer. Am J Transl Res. 2015;7(2):339–47.
Funding
University of Louisville School of Medicine Collaborative Matching Grant (HH), University of Louisville Clinical and Translational Science Pilot Grant Program Innovative Award (KMM), Melanoma Research Foundation Established Investigator Award (KMM). The University of Louisville Genomics Facility is supported by NCRR IDeA Awards INBRE-P20RR016481 and COBRE-P20RR018733, the James Graham Brown Foundation, and user fees.
Disclosures
Kelly M. McMasters reports that he is on the Board of Directors for Provectus Biopharmaceuticals and serves on the Scientific Advisory Board for Elucida Oncology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Egger, M.E., Xiao, D., Hao, H. et al. Unique Genes in Tumor-Positive Sentinel Lymph Nodes Associated with Nonsentinel Lymph Node Metastases in Melanoma. Ann Surg Oncol 25, 1296–1303 (2018). https://doi.org/10.1245/s10434-018-6377-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-018-6377-x