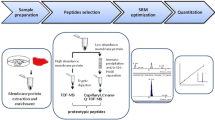
Abstract
Huge variation of drug transporter abundance was seen in the literature, making PBPK prediction difficult when transporters play a major role. Among multiple factors such as membrane fraction, digestion, and peptide selection that contributed to such variation, peptide selection is the least discussed. Herein, a strategy was established by using a small amount of purified protein standard to select a peptide with near 100% digestion efficiency for quantitation of a transporter protein MDR1. The impact of native membrane protein’s tertiary structure on the digestion efficiency of surrogate peptides of MDR1 was investigated. Peptides in more solvent accessible regions are found to be digested much more efficiently than those in large stretches of helical structures. The concentration of peptide EALDESIPPVSFWR(EAL) in the most solvent accessible linker region of MDR1 was found closest to the true protein concentration. When using EAL for MDR1 quantitation, the abundance is over 10 times higher than previously reported, indicating the importance of peptide selection for transporter quantitation. In addition, this study also proposes a screening strategy to select peptides appropriate for relative quantitation for in vitro-in vivo extrapolation in the absence of any protein standard.





Similar content being viewed by others
Change history
06 June 2018
Liling Liu was not noted as the co-first author in the original article. Buyun Chen and Liling Liu contributed equally to the article.
References
Li N, Palandra J, Nemirovskiy OV, Lai Y. LC−MS/MS mediated absolute quantification and comparison of bile salt export pump and breast cancer resistance protein in livers and hepatocytes across species. Anal Chem. 2009;81(6):2251–9.
Ji C, Tschantz WR, Pfeifer ND, Ullah M, Sadagopan N. Development of a multiplex UPLC-MRM MS method for quantification of human membrane transport proteins OATP1B1, OATP1B3 and OATP2B1 in in vitro systems and tissues. Anal Chim Acta. 2012;717:67–76.
Balogh L, Kimoto E, Chupka J, Zhang H, Lai Y. Membrane protein quantification by peptide-based mass spectrometry approaches: studies on the organic anion-transporting polypeptide family. J Proteomics Bioinform. 2012;S4:003.
Peng KW, Bacon J, Zheng M, Guo Y, Wang MZ. Ethnic variability in the expression of hepatic drug transporters: absolute quantification by an optimized targeted quantitative proteomic approach. Drug Metab Dispos. 2015;43(7):1045–55.
Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm res. 2008;25(6):1469–83.
Harwood M, Neuhoff S, Carlson G, Warhurst G, Rostami-Hodjegan A. Absolute abundance and function of intestinal drug transporters: a prerequisite for fully mechanistic in vitro–in vivo extrapolation of oral drug absorption. Biopharm Drug Dispos. 2013;34(1):2–28.
Harwood MD, Achour B, Neuhoff S, Russell MR, Carlson G, Warhurst G, et al. In vitro-in vivo extrapolation scaling factors for intestinal P-glycoprotein and breast cancer resistance protein: Part I: A cross-laboratory comparison of transporter protein abundances and relative expression factors in human intestine and Caco-2 cells. Drug Metab Dispos. 2015:dmd. 115.067371.
Vildhede A, Karlgren M, Svedberg EK, Wisniewski JR, Lai Y, Noren A, et al. Hepatic uptake of atorvastatin: influence of variability in transporter expression on uptake clearance and drug-drug interactions. Drug Metab Dispos. 2014;42(7):1210–8.
Vildhede A, Wiśniewski JR, Norén A, Karlgren M, Artursson P. Comparative proteomic analysis of human liver tissue and isolated hepatocytes with a focus on proteins determining drug exposure. J Proteome res. 2015;14(8):3305–14.
Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos. 2012;40(1):83–92.
Sakamoto A, Matsumaru T, Ishiguro N, Schaefer O, Ohtsuki S, Inoue T, et al. Reliability and robustness of simultaneous absolute quantification of drug transporters, cytochrome P450 enzymes, and Udp-glucuronosyltransferases in human liver tissue by multiplexed MRM/selected reaction monitoring mode tandem mass spectrometry with nano-liquid chromatography. J Pharm Sci. 2011;100(9):4037–43.
Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, et al. Interindividual variability in hepatic OATPs and P-glycoprotein (ABCB1) protein expression: quantification by LC-MS/MS and influence of genotype, age and sex. Drug Metab Dispos. 2013:dmd. 113.053819.
Li N, Nemirovskiy OV, Zhang Y, Yuan H, Mo J, Ji C, et al. Absolute quantification of multidrug resistance-associated protein 2 (MRP2/ABCC2) using liquid chromatography tandem mass spectrometry. Anal Biochem. 2008;380(2):211–22.
Wang L, Collins C, Kelly EJ, Chu X, Ray AS, Salphati L, et al. Transporter expression in liver tissue from subjects with alcoholic or hepatitis C cirrhosis quantified by targeted quantitative proteomics. Drug Metab Dispos 2016;44(11):1752–1758.
Deo AK, Prasad B, Balogh L, Lai Y, Unadkat JD. Interindividual variability in hepatic expression of the multidrug resistance-associated protein 2 (MRP2/ABCC2): quantification by liquid chromatography/tandem mass spectrometry. Drug Metab Dispos. 2012;40(5):852–5.
Wegler C, Gaugaz FZ, Andersson TB, Wisniewski JR, Busch D, Oswald S, et al. Protein quantification of human hepatic drug transporters and metabolizing enzymes: an inter-laboratory and methodological comparison. Drug Metab Rev (Softcover ed.). 2016;48:98–98.
Harwood MD, Russell MR, Neuhoff S, Warhurst G, Rostami-Hodjegan A. Lost in centrifugation: accounting for transporter protein losses in quantitative targeted absolute proteomics. Drug Metab Dispos. 2014;42(10):1766–72.
Bünger S, Roblick UJ, Habermann JK. Comparison of five commercial extraction kits for subsequent membrane protein profiling. Cytotechnology. 2009;61(3):153–9.
Proc JL, Kuzyk MA, Hardie DB, Yang J, Smith DS, Jackson AM, et al. A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J Proteome res. 2010;9(10):5422–37.
Zhuang X, Lu C. PBPK modeling and simulation in drug research and development. Acta Pharm sin B. 2016;6(5):430–40.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics (Oxford, England). 2010;26(7):966–8.
Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics (Oxford, England). 2014;30(6):884–6.
Pan L, Aller SG. Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics. Sci Rep. 2015;5:7880.
Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490(7421):566–9.
Hrycyna CA, Airan LE, Germann UA, Ambudkar SV, Pastan I, Gottesman MM. Structural flexibility of the linker region of human P-glycoprotein permits ATP hydrolysis and drug transport. Biochemistry. 1998;37(39):13660–73.
Scott KB, Turko IV, Phinney KW. Quantitative performance of internal standard platforms for absolute protein quantification using multiple reaction monitoring-mass spectrometry. Anal Chem. 2015;87(8):4429–35.
Waas M, Bhattacharya S, Chuppa S, Wu X, Jensen DR, Omasits U, et al. Combine and conquer: surfactants, solvents, and chaotropes for robust mass spectrometry based analyses of membrane proteins. Anal Chem. 2014;86(3):1551–9.
Chen B, Liu L, Liang X, Deng A, Dean B, Plise E, et al. Effect of surfactant, solvents and digestion conditions on digestion efficiency of drug transporter proteins by trypsin. 63rd Annual Meeting of American Society for Mass Spectrometry. 2015; St. Louis.
Lau FW, Bowie JU. A method for assessing the stability of a membrane protein. Biochemistry. 1997;36(19):5884–92.
Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta Biomembr. 2004;1666(1–2):105–17.
Nakashima H, Nakagawa Y, Makino S. Detection of the associated state of membrane proteins by polyacrylamide gradient gel electrophoresis with non-denaturing detergents application to band 3 protein from erythrocyte membranes. Biochim Biophys Acta Biomembr. 1981;643(3):509–18.
Robinson NC, Tanford C. Binding of deoxycholate, triton X-100, sodium dodecyl sulfate, and phosphatidylcholine vesicles to cytochrome b5. Biochemistry. 1975;14(2):369–78.
Siepen JA, Keevil EJ, Knight D, Hubbard SJ. Prediction of missed cleavage sites in tryptic peptides aids protein identification in proteomics. J Proteome res. 2007;6(1):399–408.
Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443(7108):180–5.
Ishihama Y, Sato T, Tabata T, Miyamoto N, Sagane K, Nagasu T, et al. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat Biotechnol. 2005;23(5):617–21.
Acknowledgements
The authors thank Dr. Kebing Yu and Dr. Donald Kirkpatrick for sharing their experience of digestion bias in shot-gun proteomics.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chen, B., Liu, L., Ho, H. et al. Strategies of Drug Transporter Quantitation by LC-MS: Importance of Peptide Selection and Digestion Efficiency. AAPS J 19, 1469–1478 (2017). https://doi.org/10.1208/s12248-017-0106-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-017-0106-4