Abstract
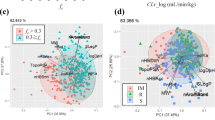
The aims were (1) to evaluate the molecular weight (MW) dependence of biliary excretion and (2) to develop quantitative structure–pharmacokinetic relationships (QSPKR) to predict biliary clearance (CLb) and percentage of administered dose excreted in bile as parent drug (PDb) in rats and humans. CLb and PDb data were collected from the literature for rats and humans. Receiver operating characteristic curve analysis was utilized to determine whether a MW threshold exists for PDb. Stepwise multiple linear regression (MLR) was used to derive QSPKR models. The predictive performance of the models was evaluated by internal validation using the leave-one-out method and external test groups. A MW threshold of 400 Da was determined for PDb for anions in rats, while 475 Da was the cutoff for anions in humans. MW thresholds were not present for cations or cations/neutral compounds in either rats or humans. The QSPKR model for human CLb showed a significant correlation (R 2 = 0.819) with good prediction performance (Q 2 = 0.722). The model was further assessed using a test group, yielding a geometric mean fold-error of 2.68. QSPKR models with significant correlation and good predictability were also developed for CLb in rats and PDb data for anions or cation/neutral compounds in rats and humans. Both CLb and PDb data were further evaluated for subsets of MRP2 or P-glycoprotein substrates, and significant relationships were derived. QSPKR models were successfully developed for biliary excretion of non-congeneric compounds in rats and humans, providing a quantitative prediction of biliary clearance of compounds.







Similar content being viewed by others
Abbreviations
- BCRP:
-
Breast cancer resistance protein
- CLb :
-
Biliary clearance
- MRP-2:
-
Multidrug resistance-associated protein 2
- MW:
-
Molecular weight
- P-gp:
-
P-glycoprotein
- PDb :
-
% of dose eliminated in the bile as parent drug
- QSPKR:
-
Quantitative structure pharmacokinetic relationship
References
Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation: physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet. 2002;41:751–90.
Ghibellini G, Leslie EM, Brouwer KL. Methods to evaluate biliary excretion of drugs in humans: an updated review. Mol Pharm. 2006;3:198–211.
Hughes RD, Millburn P, Williams RT. Biliary excretion of some diquaternary ammonium cations in the rat, guinea pig and rabbit. Biochem J. 1973;136:979–84.
Millburn P. Factors in the Biliary Excretion of Organic Compounds. In: Fishmen W, editor. Metabolic conjugation and metabolic hydrolysis. New York: Academic; 1970. p. 1–74.
Millburn P, Smith RL, Williams RT. Biliary excretion of foreign compounds: biphenyl, stilboestrol and phenolphthalein in the rat: molecular weight, polarity and metabolism as factors in biliary excretion. Biochem J. 1967;105:1275–1281.
Hughes RD, Millburn P, Williams RT. Molecular weight as a factor in the excretion of monoquaternary ammonium cations in the bile of the rat, rabbit and guinea pig. Biochem J. 1973;136:967–78.
Abou-El-Makarem MM, Millburn P, Smith RL, Williams RT. Biliary excretion of foreign compounds. Benzene and its derivatives in the rat. Biochem J. 1967;105:1269–74.
Millburn P, Smith RL, Williams RT. Biliary excretion in foreign compounds. Sulphonamide drugs in the rat. Biochem J. 1967;105:1283–7.
Hubbard RS, Butsch WL. Concentration of free sulfanilamide, sulfapyridine and sulfathiazol in material drained from human biliary tract. Proc Soc Exp Biol Med. 1941;46:484–487.
Seedorf EE, Powell WN. Experience with five orally given cholecystographic mediums. J Am Med Assoc. 1955;159:1361–2.
Alpen EL, Mandel HG, Rodwell VW, Smith PK. The metabolism of C14 carboxyl salicylic acid in the dog and in man. J Pharmacol Exp Ther. 1951;102:150–5.
Mager DE. Quantitative structure-pharmacokinetic/pharmacodynamic relationships. Adv Drug Deliv Rev. 2006;58:1326–56.
Zhou XF, Shao Q, Coburn RA, Morris ME. Quantitative structure-activity relationship and quantitative structure-pharmacokinetics relationship of 1, 4-dihydropyridines and pyridines as multidrug resistance modulators. Pharm Res. 2005;22:1989–96.
Ghafourian T, Barzegar-Jalali M, Hakimiha N, Cronin MT. Quantitative structure-pharmacokinetic relationship modelling: apparent volume of distribution. J Pharm Pharmacol. 2004;56:339–50.
Gobburu JV, Chen EP. Artificial neural networks as a novel approach to integrated pharmacokinetic-pharmacodynamic analysis. J Pharm Sci. 1996;85:505–10.
Gobburu JV, Shelver WH. Quantitative structure-pharmacokinetic relationships (QSPR) of beta blockers derived using neural networks. J Pharm Sci. 1995;84:862–5.
Herman RA, Veng-Pedersen P. Quantitative structure-pharmacokinetic relationships for systemic drug distribution kinetics not confined to a congeneric series. J Pharm Sci. 1994;83:423–8.
Mager DE, Jusko WJ. Quantitative structure-pharmacokinetic/pharmacodynamic relationships of corticosteroids in man. J Pharm Sci. 2002;91:2441–51.
Ng C, Xiao Y, Putnam W, Lum B, Tropsha A. Quantitative structure-pharmacokinetic parameters relationships (QSPKR) analysis of antimicrobial agents in humans using simulated annealing k-nearest-neighbor and partial least-square analysis methods. J Pharm Sci. 2004;93:2535–44.
Van der Graaf PH, Nilsson J, Van Schaick EA, Danhof M. Multivariate quantitative structure-pharmacokinetic relationships (QSPKR) analysis of adenosine A1 receptor agonists in rat. J Pharm Sci. 1999;88:306–12.
Andrews CW, Bennett L, Yu LX. Predicting human oral bioavailability of a compound: development of a novel quantitative structure-bioavailability relationship. Pharm Res. 2000;17:639–44.
Martin PD, Warwick MJ, Dane AL, Brindley C, Short T. Absolute oral bioavailability of rosuvastatin in healthy white adult male volunteers. Clin Ther. 2003;25:2553–63.
Martin PD, Warwick MJ, Dane AL, Hill SJ, Giles PB, Phillips PJ, et al. Metabolism, excretion, and pharmacokinetics of rosuvastatin in healthy adult male volunteers. Clin Ther. 2003;25:2822–35.
Bewick V, Cheek L, Ball J. Statistics review 13: receiver operating characteristic curves. Crit Care. 2004;8:508–12.
Galley HF. Editorial II: Solid as a ROC. Br J Anaesth. 2004;93:623–6.
Harter M, Reuter K, Gross-Hardt K, Bengel J. Screening for anxiety, depressive and somatoform disorders in rehabilitation–validity of HADS and GHQ-12 in patients with musculoskeletal disease. Disabil Rehabil. 2001;23:737–44.
Strik JJ, Honig A, Lousberg R, Denollet J. Sensitivity and specificity of observer and self-report questionnaires in major and minor depression following myocardial infarction. Psychosomatics. 2001;42:423–8.
Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36.
Draper NH, Smith H. Applied regression analysis. 3rd ed. Hoboken: Wiley; 1998.
Parrott N, Paquereau N, Coassolo P, Lave T. An evaluation of the utility of physiologically based models of pharmacokinetics in early drug discovery. J Pharm Sci. 2005;94:2327–43.
Giuliano C, Fiore F, Di Marco A, Padron Velazquez J, Bishop A, Bonelli F, et al. Preclinical pharmacokinetics and metabolism of a potent non-nucleoside inhibitor of the hepatitis C virus NS5B polymerase. Xenobiotica. 2005;35:1035–54.
Rollins DE, Klaassen CD. Biliary excretion of drugs in man. Clin Pharmacokinet. 1979;4:368–79.
Hirom PC, Millburn P, Smith RL, Williams RT. Species variations in the threshold molecular-weight factor for the biliary excretion of organic anions. Biochem J. 1972;129:1071–7.
Fouchecourt MO, Beliveau M, Krishnan K. Quantitative structure-pharmacokinetic relationship modelling. Sci Total Environ. 2001;274:125–35.
Ueda K, Taguchi Y, Morishima M. How does P-glycoprotein recognize its substrates? Semin Cancer Biol. 1997;8:151–9.
Pearce HL, Safa AR, Bach NJ, Winter MA, Cirtain MC, Beck WT. Essential features of the P-glycoprotein pharmacophore as defined by a series of reserpine analogs that modulate multidrug resistance. Proc Natl Acad Sci U S A. 1989;86:5128–32.
Brauer RW. Mechanisms of bile secretion. J Am Med Assoc. 1959;169:1462–6.
Gombar VK, Polli JW, Humphreys JE, Wring SA, Serabjit-Singh CS. Predicting P-glycoprotein substrates by a quantitative structure-activity relationship model. J Pharm Sci. 2004;93:957–68.
Ha SN, Hochman J, Sheridan RP. Mini review on molecular modeling of P-glycoprotein (Pgp). Curr Top Med Chem. 2007;7:1525–9.
Zhang S, Yang X, Coburn RA, Morris ME. Structure activity relationships and quantitative structure activity relationships for the flavonoid-mediated inhibition of breast cancer resistance protein. Biochem Pharmacol. 2005;70:627–39.
van Zanden JJ, Wortelboer HM, Bijlsma S, Punt A, Usta M, Bladeren PJ, et al. Quantitative structure activity relationship studies on the flavonoid mediated inhibition of multidrug resistance proteins 1 and 2. Biochem Pharmacol. 2005;69:699–708.
Hinderling PH, Schmidlin O, Seydel JK. Quantitative relationships between structure and pharmacokinetics of beta-adrenoceptor blocking agents in man. J Pharmacokinet Biopharm. 1984;12:263–87.
Smith DA, van de Waterbeemd H, Walker DK, Mannhold R, Kubinyi H, Timmerman H. Pharmacokinetics and metabolism in drug design. Weinheim: Wiley; 2001. p. 59–66.
Obach RS, Lombardo F, Waters NJ. Trend analysis of a database of intravenous pharmacokinetic parameters in humans for 670 drug compounds. Drug Metab Dispos. 2008;36:1385–405.
Smith RL. The excretory function of bile. London: Chapman & Hall; 1973. p. 16–34.
Acknowledgements
This work was supported in part by Pfizer Inc. We thank Dr. Lisa J. Benincosa from the Groton/New London Laboratories at Pfizer Inc. for her support and suggestions regarding this research. We thank Dr. Xueya Cai from Division of Biostatistics of Indiana University for her help with receiver operating characteristic curves analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
The complete dataset of compounds with biliary clearance and percentage of dose eliminated in bile data, in both rats and humans, along with information on administration route, collection period of samples, analytical method, MW of the compounds, charge status of the compounds under physiological conditions (pH = 7.4) and transporters known to mediate efflux processes of the drugs, are available as Excel files. A table containing the QSARis© descriptors is also present. This information can be found at http://www.buffalo.edu/~memorris
Rights and permissions
About this article
Cite this article
Yang, X., Gandhi, Y.A., Duignan, D.B. et al. Prediction of Biliary Excretion in Rats and Humans Using Molecular Weight and Quantitative Structure–Pharmacokinetic Relationships. AAPS J 11, 511–525 (2009). https://doi.org/10.1208/s12248-009-9124-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-009-9124-1