Abstract
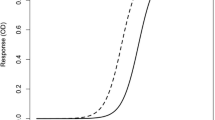
Calibration curves for ligand binding assays are generally characterized by a nonlinear relationship between the mean response and the analyte concentration. Typically, the response exhibits a sigmoidal relationship with concentration. The currently accepted reference model for these calibration curves is the 4-parameter logistic (4-PL) model, which optimizes accuracy and precision over the maximum usable calibration range. Incorporation of weighting into the model requires additional effort but generally results in improved calibration curve performance. For calibration curves with some asymmetry, introduction of a fifth parameter (5-PL) may further improve the goodness of fit of the experimental data to the algorithm. Alternative models should be used with caution and with knowledge of the accuracy and precision performance of the model across the entire calibration range, but particularly at upper and lower analyte concentration areas, where the 4-and 5-PL algorithms generally outperform alternative models. Several assay design parameters, such as placement of calibrator concentrations across the selected range and assay layout on multiwell plates, should be considered, to enable optimal application of the 4- or 5-PL model. The fit of the experimental data to the model should be evaluated by assessment of agreement of nominal and model-predicted data for calibrators.
Similar content being viewed by others
References
2006 report, Medicines in development, biotechnology. PhRMA Website. 2006; Available at: http://www.phrma.org/files/Biotech%202006.pdf. Accessed January 14, 2007.
Findlay JWA, Smith WC, Lee JW, et al. Validation of immunoassays for bioanalysis: a pharmaceutical industry perspective.J Pharm Biomed Anal. 2000;21:1249–1273.
DeSilva B, Smith W, Weiner R, et al. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules.Pharm Res. 2003;20:1885–1900.
Shah VP, Midha KK, Dighe S, et al. Analytical methods validation: bioavailability, bioequivalence and pharmacokinetic studies.Pharm Res. 1992;9:588–592.
Miller KJ, Bowsher RR, Celniker A, et al. Workshop on bioanalytical methods validation for macromolecules: summary report.Pharm Res. 2001;18:1373–1383.
Shah VP, Midha KK, Findlay JWA, et al. Bioanalytical method validation—a revisit with a decade of progress.Pharm Res. 2000;17:1551–1557.
Vishwanathan CT Bansal S, Booth B, et al. Quantitative bioanalytical methods validation and implementation best practices for chromatographic and ligand binding assays.AAPS J [serial online]. 2007;9:E30-E42.
Food and Drug Administration.Guidance for Industry: Bioanalytical. Method Validation. Rockville, MD: US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research; 2001.
Rodbard D, Frazier GR. Statistical analysis of radioligand assay data.Methods Enzymol. 1975;37:3–22.
Haven MC, Orsulak PJ, Arnold LL, et al. Data-reduction methods for immunoradiometric assays of thyrotropin compared.Clin Chem. 1987;33:1207–1210.
Dudley RA, Edwards P, Ekins RP, et al. Guidelines for immunoassay data processing.Clin Chem. 1985;31:1264–1271.
Gottschalk PG, Dunn JR. The five-parameter logistic: a characterization and comparison with the four-parameter logistic.Anal Biochem. 2005;343:54–65.
Box GEP, Hunter WG, Hunter JS, eds.Statistics for Experimenters. New York, NY: John Wiley & Sons: 1978.
Finney DJ, Phillips P. The form and estimation of a variance function, with particular reference to radioimmunoassay.Appl Stat. 1977;26:312–320.
Finney DJ, ed.Statistical Methods in Biological Assay. 3rd ed. London, UK: Charles Griffith; 1978.
Carroll RJ, Ruppert D, eds.Transformation and Weighting in Regression. London, UK: Chapman Hall; 1988.
Rocke DM, Jones G. Optimal design for ELISA and other forms of immunoassay.Technometrics. 1997;39:162–170.
Karpinski KF. Optimality assessment in the enzyme-linked immunosorbent assay (ELISA).Biometrics. 1990;46:381–390.
Singer R, Lansky DM, Hauck WW. Bioassay glossary.Pharmacopeial Forum. 2006;32:1359–1365.
Karnes HT, March C. Calibration and validation of linearity in chromatographic biopharmaceutical analysis.J Pharm Biomed Anal. 1991;9:911–918.
Smith WC, Sittampalam GS. Conceptual and statistical issues in the validation of analytic dilution assays for pharmaceutical applications.J Biopharm Stat. 1998;8:509–532.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published: June 29, 2007
Rights and permissions
About this article
Cite this article
Findlay, J.W.A., Dillard, R.F. Appropriate calibration curve fitting in ligand binding assays. AAPS J 9, 29 (2007). https://doi.org/10.1208/aapsj0902029
Received:
Accepted:
DOI: https://doi.org/10.1208/aapsj0902029