Abstract
Since its discovery more than 110 years ago, Valsa canker has emerged as a devastating disease of apple in East Asia. However, our understanding of this disease, particularly the identity of the causative agents, has been in a state of confusion. Here we provide a synopsis for the current understanding of Valsa canker and the taxonomy of its causal agents. We highlight the major changes concerning the identity of pathogens and the conflicting viewpoints in moving to “One Fungus = One Name” system for this group of fungal species. We compiled a list of 21 Cytospora species associated with Malus hosts worldwide and curated 12 of them with rDNA-ITS sequences. The inadequacy of rDNA-ITS in discriminating Cytospora species suggests that additional molecular markers, more intraspecific samples and robust methods are required to achieve reliable species recognition.
Similar content being viewed by others
Background
Apple (Malus domestica Borkh) is one of the most widely planted and nutritionally important fruit crops in the world (Cornille et al. 2014; Duan et al. 2017). At one time nearly each area had its own local apple cultivars (Janick et al. 1996). As of 2018, the overall apple plantation area was close to 5 million ha and the total apple production exceeded 86 million ton, ranking the third in worldwide fruit production (http://www.fao.org/faostat/). The nutritional value of apple makes this fruit particularly beneficial to human health as being highlighted by a famous aphorism: “An apple a day keeps the doctor away”. With the increase of people’s income and public awareness for balanced nutrition, apple consumption continues to grow beyond its global production capacity. In the meanwhile, apple production, and global food security in general, faces a tremendous threat from fungal diseases (Fisher et al. 2012). It has been estimated that 12–25% of annual apple harvest was lost due to abiotic and biotic stresses, including those caused by fungal diseases (Forte et al. 2002).
Valsa canker has emerged as a global threat to apple industry (CABI and EPPO 2005; EPPO 2020), and is particularly destructive in East Asia (Togashi 1925; Uhm and Sohn 1995; Abe et al. 2007; Wang et al. 2011). Its causative fungus, Valsa mali (Ideta 1909; Tanaka 1919; Togashi 1925; Tai 1979), was once synonymized with V. ceratosperma (Kobayashi 1970), and has recently been resurrected (Adams et al. 2005; Wang et al. 2011, 2014). It is critically important to have pathogens be settled in a stable taxonomy with clearly defined species boundary, because species are the fundamental unit not only in communicating the related knowledge of the pathogen but also for linking information regarding pathogen biology, host range, distribution, and potential risk (Crous et al. 2015). Well-defined species allow practitioners to reliably identify the pathogens, accurately diagnose the diseases and effectively develop the corresponding management strategies. Unfortunately, this has not been the case in the near-120-year history of the pathogens causing apple Valsa canker. Besides, our knowledge on how to identify species has undergone revolutionary advances, which in turn has incurred nomenclatural changes throughout time (Rossman and Palm-Hernández 2008). We have seen remarkable changes in taxonomy of Valsa canker pathogens, particularly with the advent of DNA-based species identification technique and our acceptance of the “one fungus one name” principle, following which the genus name Cytospora is used instead of Valsa (Rossman et al. 2015).
The aim of the present review is to provide a synopsis of the current state of our understanding of Valsa canker disease and the taxonomy of its causal agents in East Asia, highlighting those areas where substantial changes have been taken place and conflicting viewpoints have been emerged, and to revisit the fungal species associated with Malus hosts in the new taxonomy framework based on the available DNA sequence data, placing the needs of the practitioners, especially plant pathologists, at the forefront.
Overview of Valsa canker disease of apple
Discovery
Valsa canker was first discovered in Japan at the beginning of the twentieth century (Ideta 1909; Tanaka 1919). It has spread in apple orchards since the introduction of the American apple varieties and caused serious cankers on trunks and branches (Togashi 1925). Although the causal fungus was described under the name of Valsa mali Miyabe et Yamada, the disease had been confused with the fire-blight caused by bacteria Bacillus amylovorus before the causative agent was verified through inoculation experiments (Tanaka 1919; Togashi 1925). This fungal species was also listed as the causal pathogen of canker disease of apple in China, Korea and Far East of Russia (Tai 1979; Vasilyeva and Kim 2000; Wang et al. 2011). In North America, a canker disease on the main trunks of young apple trees capable of causing much damage was first reported from Illinois of the USA in 1919. The fungus associated with the canker was not formally identified but was thought to be “agreed well with Cytospora of Valsa leucostoma” (Stevens 1919). In the following years, Valsa cankers were found to be common on apple trees in New Mexico (Leonian 1921). In Europe, a large number of cases of canker and die-back of apple trees were reported lately and the pathogen was identified as Valsa ambiens (Ogilvie 1933). More recently, another species, Leucostoma cincta, was responsible to outbreaks of canker disease of apple in North America (Proffer and Jones 1989; Brown-Rytlewski and McManus 2000). In the old taxonomic system of fungi, asexual morphs of both Valsa and Leucostoma were included in the genus Cytospora. All above historical clues indicate that the underlying pathogen species for the canker disease of apple might differ from region to region.
Common names of the disease
Traditionally, the name of a specific disease is unique and often directly related to its causing agent. However, there are cases in which different names were used for the same disease, for example, Valsa canker, Leucostoma canker, and Cytospora canker were indeed all pointing to apple Valsa canker. This is a source of confusion for plant pathologists. Besides, some even more ambiguous names, such as “Apple Canker” and “Perennial Canker” were also used in literatures referring to apple Valsa canker. Cautions should be taken when dealing with these diseases in literatures. A disease called “Apple Canker” was caused by Sphaeropsis malorum (Paddock 1898) and appeared more than 120 years ago, which is now referred to as apple black rot. In fact, the name “Apple Canker” is more commonly referred to the disease caused by Neonectria ditissima. Moreover, as the symptom of canker can be caused by both bacteria and fungi (Turechek 2004), pathogen information conveyed by disease name “Apple Canker” is scant. Similarly, the disease initially called “Perennial Canker” of apple trees was not caused by fungi of Valsa/Cytospora. It was first used by Zeller and Childs (1925) to refer to a fungal disease caused by Gloeosporium perennans. Both of these names are not informative and prone to confusion or misunderstanding, thus should be avoided when referring to the canker disease caused by Valsa/Cytospora species.
As far as the perennial canker disease of apple in East Asia is concerned, “apple Valsa canker” appears to be an informative and specific name. This name directly hinges on the fact that the causal pathogen of the disease was initially described based on its sexual (Valsa) form. Its asexual name in Cytospora was assigned lately but was never attached to the disease name (Kobayashi 1970; Vasilyeva and Kim 2000). This is due to the fact that the sexual morph was preferred in traditional taxonomic treatments and the morphological classification of its asexual form has been a persistent confusion. In fact, the canker disease of apple caused by the fungus Valsa mali (even other synonym was used) was exclusive referred to as apple Valsa canker irrespective of whether sexual or asexual forms were investigated. In addition, with recent acceptance of using genus name Cytospora instead of Valsa (Rossman et al. 2015), the common disease name “apple Valsa canker” becomes even more unique, which minimizes confusion but maximizes information content at the same time.
Symptom, infection and damage
Apple Valsa canker is characterized by elongated cankers on trunks and scaffold limbs or dieback of twigs (Suzaki 2008). The disease first appears on the branch surface as brownish oblong spots with irregular edges (Tanaka 1919). The infected tissues then gradually dry out, slightly sunken, more or less darkened, and finally form localized cankers. The canker develops rapidly between spring and early summer, and then slowly during the late summer and in the winter (Abe et al. 2007). With perennial development of cankers, they girdle twigs, branches and trunks, which lead to desiccation and death of the distal part, even the entire tree.
The fungus infects apple trees through natural bark crevices and wounds due to adverse climatic conditions or pruning and other mechanical injuries. Infected hyphae develop inter- and intra-cellularly and penetrate extensively into the phloem and xylem of trunks and main crotches (Tamura and Saito 1982; Ke et al. 2013), leading to perennial infections which cannot be effectively controlled through chemical treatments or surgical removes (Abe et al. 2007). Moreover, recent studies have indicated that infection can be asymptomatic (Zang et al. 2012; Zhang et al. 2018), and latent infections in seeds, seedlings and twigs provide a potential inoculum source for the disease (Meng et al. 2019). Although numerous management approaches, including physical, chemical and biological methods, have been implemented to limit the disease, apple Valsa canker remains prevalent in many apple production areas (Peng et al. 2016).
Since its discovery, apple Valsa canker has emerged as a global problem to apple industry, seriously impeding apple production, especially in East Asian countries of China (Chen et al. 1987; Wang et al. 2005; Cao et al. 2009; Wang et al. 2011), Korea (Uhm and Sohn 1995; Lee et al. 2006), and Japan (Sawamura et al. 1990; Janick et al. 1996; Abe et al. 2007), where nearly half of the world total apple yield was produced (http://www.fao.org/faostat/). All commercially important apple varieties are susceptible to Valsa canker (Bessho et al. 1994). With the expanding requirements for high yield and commercial quality, nowadays a few apple cultivars dominate the major apple planting regions. Such a large scale homogenization of germplasm exposes modern apple orchards at high risk of outbreaks for Valsa canker. In some regions, few orchards are free from its damaging effects; the average incidence ranges from 10 to 55% in the major apple growth regions of China (Wang et al. 2005; Cao et al. 2009; Li et al. 2013).
Discerning apple Valsa canker pathogens in East Asia
Species status of Valsa mali
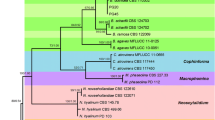
The pathogen of apple Valsa canker was initially recognized as a new species, Valsa mali Miyabe et Yamada (Ideta 1909). Based on the morphological and cultural characteristics, Kobayashi (1970) concluded that V. mali was identical to V. ceratosperma (Tode ex Fries) Maire and recognized its anamorph as Cytospora rosarum but not the commonly used name C. sacculus (Spielman 1985; Adams et al. 2005). Kobayashi’s (1970) classification of asexual state of V. mali was not adopted. Since then, both V. mali and V. ceratosperma (and/or C. sacculus) have been used to refer to the causative agents of apple Valsa canker in East Asia (Wang et al. 2011). However, V. ceratosperma is morphologically heterogeneous, including both small-spored and large-spored specimens that infect a wide array of host plants (Kobayashi 1970; Spielman 1985; Old et al. 1991). Further evidence from both morphological characterization and molecular phylogenetic analysis supports that V. mali is an independent species. Stable morphological features differentiating V. mali from typical strains of V. ceratosperma have been listed by Spielman (1985) and confirmed by later re-description (Lu 1992; Vasilyeva and Kim 2000; Wang et al. 2011). Recent phylogenetic analysis of rDNA-ITS sequences revealed that V. ceratosperma was not a monophyletic clade but represented a heterogeneous species complex (Fig. 1), including at least three deeply diverged lineages (Adams et al. 2005). These lineages were recognized as distinct species, each parasitizing a narrow spectrum of host plants (Adams et al. 2005; Wang et al. 2011). A lineage composed of a strain from apple in Japan was referred to as “V. ceratosperma sensu Kobayashi” by Adams et al. (2005). In a further analysis, the ITS sequence of this strain clustered with apple and pear strains in China and also with that of the neotype of V. mali (Vasilyeva and Kim 2000), forming a strongly supported monophyletic clade (Wang et al. 2011). The clade of V. mali is more closely related to V. malicola than to other members in the V. ceratosperma complex (Fig. 1). These evidences support the view that V. mali from East Asia represents a morphologically distinct and genetically divergent clade as well as an evolutionarily independent lineage distinct from other members of the V. ceratosperma complex (Wang et al. 2011).
Phylogenetic relationship among Valsa ceratosperma species complex reconstructed from ITS sequences. V. ceratosperma sensu lato is not a monophyletic group with respect to V. malicola. The lineage of V. ceratosperma sensu Kobayashi (Adams et al. 2005) was recognized as V. mali (Wang et al. 2011, 2014) within which a cryptic species, V. pyri, was further recognized based on multi-gene phylogenetic analysis (Wang et al. 2014)
Confusion about the species status of V. mali was introduced by a strain (CBS367.29, GenBank No. AF191186) from Japan (Adams et al. 2002). Although this strain was labeled as “Valsa mali Miyabe & Yamada”, it was indeed part of the Leucostoma persoonii group (Adams et al. 2002, 2005) and did not belong to Valsa mali Miyabe et Yamada which was re-described and re-typified by Vasilyeva and Kim (2000). Analysis of rDNA-ITS sequence of the ex-type of Vasilyeva and Kim’s strain and the strain labeled as “V. ceratosperma sensu Kobayashi” by Adams et al. (2005) confirmed that they belonged to the same species (Wang et al. 2011).
Valsa pyri: a cryptic species within V. mali
Previous researches revealed that there was cryptic divergence within V. mali (Lu 1992; Wang et al. 2014). Isozyme electrophoresis revealed that strains infecting pear were clearly differentiated from strains infecting apple although they were morphologically very similar (Lu 1992). The strains from pear were recognized as an independent variety, V. mali var. pyri, to distinguish it from the apple strains, V. mali var. mali (Lu 1992). This result was confirmed by recent phylogenetic analysis of rDNA-ITS sequences, which revealed strongly supported reciprocal monophyletic clades for both varieties with a p-distance of 1.4% (Wang et al. 2011). Additional evidence has accumulated to support that these two varieties are actually distinct species.
Analyzing DNA sequences of rDNA-ITS, β-tubulin and EF1α independently, followed by delineating species based on genealogical concordance phylogenetic species recognition (GCPSR) (Taylor et al. 2000), and delimitating species using all loci together with the multispecies coalescence model (Heled and Drummond 2010; Yang and Rannala 2010), consistently support that these two varieties represent two independent species, V. pyri and V. mali, respectively (Wang et al. 2014). Although their genetic divergence in rDNA-ITS appears low (1.4%), their sequence divergences in β-tubulin and EF1α are high, with a mean pairwise distance of 8.0 and 5.1%, respectively (Fig. 2). Secondly, V. pyri and V. mali show distinct cultural characteristics. Colonies of V. pyri remained milky white throughout the observation period (with rare exception), but those of V. mali changed from white to light brown during the same period of time (Lu 1992; Wang et al. 2011). In addition, the temperature range for normal fungal growth differs between species. At 37 °C, V. pyri grew normally on PDA medium whereas V. mali did not (Wang et al. 2011). Third, V. pyri and V. mali manifest distinct patterns of pathogenicity. Strains of V. mali were found infecting species of Malus, Prunus, and Crataegus (Vasilyeva and Kim 2000; Wang et al. 2011; Fan et al. 2020), while strains of V. pyri were only isolated from Malus and Pyrus. In cross-inoculation tests, V. mali was more aggressive on apple than on pear, while the reverse was true for V. pyri regardless of the source of host plant (Zang et al. 2007; Wang et al. 2011). Finally, the results of whole genome sequencing revealed that two species differ sharply in genome content. The genome size is 44.7 Mb for V. mali and 35.7 Mb for V. pyri. The genome of V. pyri possesses far fewer repetitive elements. Although both species possess genes that produce proteins associated with pectin decomposition, they show distinct variations in many secondary metabolism gene clusters (Yin et al. 2015). Such a large genomic difference was accumulated during more than 5 million years of independent evolution of the two species (Wang et al. 2014).
Genetic divergence between Valsa mali and Valsa pyri. The phylogenetic relationship is illustrated by a network constructed from the concatenated alignments of DNA sequences for three loci: ITS, β-tubulin and EF1α. The means and standard errors of the p-distances calculated from each locus are shown in the inserted table on the branch separating the species. This figure was modified from Wang et al. (2014)
Moving to “One Fungus = One Name” for apple Valsa canker pathogens
Like many other fungal species, pathogens of Valsa canker are traditionally recognized based on morphological features. Because the asexual forms (anamorphs) and sexual forms (teleomorphs) can develop independently and their relationship is often uncertainly judged from morphology, these fungi often have dual names describing asexual and sexual forms, respectively. With the application of molecular methods, particularly those based on DNA sequences, it can be confirmed that these separate forms are indeed a single species (e.g. Cannon and Kirk 2000; Taylor 2011; Wang et al. 2011; Crous et al. 2016), and thus applying separate names to different forms of the same species becomes redundant (Hawksworth et al. 2011; de Hoog et al. 2015). For this reason, recent rule in the International Code of Nomenclature for algae, fungi, and plants (ICN) is advancing a move towards “One Fungus = One Name” nomenclatural system (McNeill et al. 2012). Changes in ICN rule have a particularly consequential effect on fungi causing Valsa canker. The asexual forms of this disease were recognized as Cytospora, which also includes sexual counterparts of other related genera Leucocytospora, Leucostoma, Valsella, and Valseutypella besides Valsa (Adams et al. 2005; Rossman et al. 2015). Before July 2011 when the “One Fungus = One Name” rule came into force (Hawksworth et al. 2011), both sexual names and asexual names were used for this group of fungi and in some cases multiple combinations of sexual and asexual names referring to the same species were available in literatures. During the initial move to “One Fungus = One Name” according to the earlier version of ICN (McNeill et al. 2012), the teleomorph-typified species names were preferred. Thus the sexual generic name Valsa was preferred to asexual generic name Cytospora. However, the article for the preference of teleomorph-typified genus name was removed from the newest version of ICN (Turland et al. 2017, 2018) which requires researchers to use the genus name Cytospora 1818 instead of Valsa 1825 according exclusively to the principle of priority (Rossman et al. 2015). As a consequence, the genus name for all species formerly recognized under Valsa need to be changed into Cytospora. To date (as of May 31, 2020), 406 Cytospora names and 482 Valsa names have been recorded in the database of Species Fungorum (http://www.speciesfungorum.org/). Even more species name epithets have been listed in the database of Index Fungorum (http://www.indexfungorum.org/). For many of these species, the validity needs to be rigorously tested and the link between Cytospora and Valsa names still awaits to be reliably established before they can be finally moved into an “One Fungus = One Name” system. Kirk et al. (2008) only listed approximately 110 accepted Cytospora species, while all other species names were considered synonyms of previously described taxa or non-Cytospora species. It is unclear how many species still remain valid under the “One Fungus = One Name” taxonomic framework. At the early stage of nomenclatural move confusions are inevitable, but in the long run the move to “One Fungus = One Name” will lead to more stable and meaningful taxonomy of these fungal species.
According to the newest outline for classification of fungi, the species causing apple Valsa canker belongs to phylum Ascomycota, class Sordariomycetes, order Diaporthales, family Cytosporaceae (= Valsaceae), and genus Cytospora (Wijayawardene et al. 2020). While linking asexual and sexual forms of the same species becomes straightforward with the aids of DNA sequences, an issue of “One Name = Which Name” has emerged from the existing species since decisions must be made with regard to previous names for both asexual and sexual forms as well as their synonyms (Hawksworth 2012). Such a move is not straightforward and confusions have already been introduced.
From Valsa mali to Cytospora mali
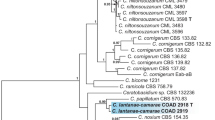
Following the ICN rule, the species name Valsa mali should be changed into Cytospora mali. However, the epithet C. mali has already been used by Grove (1935) to refer to a strain on apple from USA which was originally described by Stevens (1919). Lines of evidence suggest that V. mali Miyabe et Yamada and C. mali Grove might represent different species. Firstly, Stevens (1919) thought the isolate treated as C. mali by Grove “agrees well with the Cytospora of Valsa leucostoma”. Secondly, conidia size of V. mali (3–6 × 0.5–1.5 μm, Kobayashi 1970) was clearly different from that of C. mali (7 × 1.6 μm, Grove 1935). Lastly, in phylogenetic analysis of rDNA-ITS sequences, no USA isolate was clustered within the clade recognized as V. mali (Fig. 3). Grove’s species name was included in MycoBank (http://www.mycobank.org/) but was not included in Index of Plant Diseases in the United States (USDA 1960).
Cytospora species on Malus hosts. Neighbor-joining phylogeny constructed using ITS sequences with the Tamura-Nei model. Sequences for each species record from apple were used as entries and blasted in GenBank with a coverage score of > 97% and an identity score of 98.5% that is equivalent to the 1.5% threshold used by the UNITE database for molecular identification of fungi (Nilsson et al. 2019). This threshold is conservative for fungi (Vu et al. 2019) and the blast hits are much more inclusive. Only the representative sequences with information on host and geographic origins were included. The phylogenetic tree was linearized for clarity
Recently, Fan et al. (2020) synonymized V. mali Miyabe et Yamada with C. mali Grove without critical justification in a treatment of Cytospora species from China. The alignment of rDNA-ITS sequences of isolates referred to as C. mali Grove by Fan et al. (2020) and other Cytospora species from apple showed that they have high similarity and clustered together to form a strongly supported monophyletic clade (Fig. 3 and Additional file 1: Figure S1). This result indicates that C. mali Grove referred to by Fan et al. (2020) is actually V. mali. Even assuming that Grove’s species is the same species as V. mali, it was preceded by V. mali. The name V. mali first appeared in a list of fruit disease compiled by Sapporo Agricultural College during 1903–1904 and was formally described by Takahashi and Okamoto in 1908 (Tanaka 1919). Ideta (1909) gave a more detailed account of the fungus together with the original drawing of Yamada in the following year. A full English description for this species was also available (Tanaka 1919) before Kobayashi (1970) synonymized V. mali with V. ceratosperma. Vasilyeva and Kim (2000) provided a thorough re-description based on the assigned neotype from cultivated Malus sp. All above evidence suggests that C. mali Grove is not a proper name or an epithet for this species and the pathogen for apple Valsa canker in East Asia should be properly referred to as C. mali (Miyabe et Yamada).
From Valsa pyri to Cytospora pyri
Confusion also occurs when moving from Valsa pyri to Cytospora pyri. A species epithet, C. pyri Fuckel 1860 (Cytispora pyri, in its original spelling), has already been listed in Index Fungorum. Fuckel’s species is now treated as a synonym of Discular pyri, a species of family Gnomoniaceae but not family Valsaceae (von Höhnel 1926). In addition, a recent provisional treatment of a phylogenetic clade corresponding to V. pyri as C. leucosperma by Fan et al. (2020) incurs a new confusion. V. pyri was clearly distinct from C. leucosperma (syn. V. ambiens) as described by Adams et al. (2005). Although it is hard to link Spielman’s (1985) lectotype to the isolates from different studies without molecular data, the record of hosts also suggests that the isolates studied by Fan et al. (2020) were V. pyri rather than C. leucosperma. No specimens that Spielman (1985) identified as C. leucosperma was from plant family of Rosaceae, while all materials examined by Fan et al. (2020) were collected from Pyrus bretschneideri, a member of Rosaceae, from northwest China where V. pyri has been reported (Wang et al. 2011). In summary, Fuckel’s (1860) species of C. pyri is not the same species as V. pyri and the name is now invalid, and Fan et al.’s (2020) species of C. leucosperma is actually V. pyri. Thus, it is suitable to refer the pathogen species causing perennial canker disease on both apple and pear as C. pyri (sensu V. mali var. pyri Lu 1992) in accordance with “One Fungus = One Name” rule of new ICN.
Cytospora species associated with Malus hosts
As many as 21 species of Cytospora and their sexual allies have been reported from Malus worldwide, 11 of which occur in East Asia (Table 1). Most of these species have not been systematically investigated and the taxonomic status of only a few of them has been validated through molecular approaches. We re-evaluated status of these species here based on rDNA-ITS sequences, a universal marker that has been widely used for molecular identification of fungi, i.e. DNA barcoding (Schoch et al. 2012). ITS sequences were available for 12 of the 21 listed Cytospora species from Malus plants. We blasted the sequences of apple isolates in GenBank and downloaded the homologous hits that have coverage scores of > 97% and identity scores of 98.5%. This identity score is equivalent to the 1.5% threshold used by the UNITE database (Nilsson et al. 2019), and is conservative in molecular identification of fungi (Vu et al. 2019). Only the representative sequences with information of host and geographic origins were included. For the identical sequences, we kept only one representative from each host plant and each geographic region. The phylogeny constructed using these ITS sequences revealed some interesting results for Cytospora species infecting Malus host (Fig. 3).
First, not all species of Cytospora can be distinguished based on ITS sequences. In consistence with earlier studies, a recently described species C. parasitica was indistinguishable from C. shulzeri (Ariyawansa et al. 2015; Norphanphoun et al. 2017). C. parasitica falls within diversity of C. shulzeri (Fig. 3). The result of reciprocal monophyly for these two species in a recent study (Ma et al. 2018) was due to poor sampling of intraspecific diversity, in which only a sub-lineage of C. shulzeri was included. Similarly, C. mali-spectabilis clustered within the clade of C. nivea in the phylogenetic tree and more strains are needed to validate its taxonomic status. On the contrary, two species, C. cincta and C. leucostoma, manifest high genetic heterogeneity. It remains to be determined whether additional cryptic species exist within the named species. Second, no Cytospora species occurred exclusively on Malus. All Cytospora species found on Malus also infect other plants, no matter how similar their rDNA-ITS sequences are. Indeed, there are cases in which the haplotype with the same rDNA-ITS sequences was isolated from different host plant genera, e.g., Malus and Pyrus (Wang et al. 2011). Third, Cytospora species infecting Malus are geographically widespread. All species occurred in more than one country except two recently described ones that were only known by type localities. Fourth, the distribution of coexisting species and prevalence of the pathogens vary from region to region (Wang et al. 2011; Yin et al. 2016). Below we summarize these 12 molecularly curated species one by one following the order they appear in the phylogenetic tree (Fig. 3).
Cytospora mali (Miyabe et Yamada, 1909)
This species is the major pathogen responsible for apple Valsa canker in East Asia (Kobayashi 1970; Vasilyeva and Kim 2000; Adams et al. 2005; Wang et al. 2011). It was known for its sexual form and subjected to synonymization. Thus several asexual names or combinations of sexual and asexual names have been used in literature. Tracing back through clues from morphology, geography and host, the species concept discussed here is consistent with the original description of V. mali (Miyabe and Yamada 1915; Vasilyeva and Kim 2000), V. ceratosperma sensu Kobayashi (1970), and V. mali var. mali sensu Wang et al. (2011), but is not with C. mali Grove (1935). Tracing through ITS sequence data, the known distribution of this species includes China, Japan, South Korea and Far East of Russia (Adams et al. 2005; Wang et al. 2011; Yin et al. 2016). Its occurrence in other regions of world remains to be verified. C. mali was predominant among isolates from apple in Shaanxi Province, China (Wang et al. 2011). Other hosts of C. mali include species of Crataegnus, Morus and Prunus (Fan et al. 2020).
Cytospora pyri (Lu 1992)
This species was formerly thought as a variety of C. mali (Lu 1992; Wang et al. 2011; Zhou et al. 2013), and was recently delimited as full species based on multilocus phylogenetic analysis (Wang et al. 2014). It was commonly found on Pyrus hosts and responsible for canker disease of pear (Lu 1992; Wang et al. 2011; Yin et al. 2016). Geographic distribution of C. pyri still remains to be fully defined and is currently known in China and Italy (Wang et al. 2011). Occurrence of this species on Malus tree was generally rare but disease incidence it caused may be high in some orchards, even exceeded that of C. mali (Yin et al. 2016). However, experimental inoculation tests indicated that it was less virulent on apple than on pear (Wang et al. 2011).
Cytospora schulzeri Sacc. & P. Syd. 1899
The telemorph of this species was formerly known as Valsa malicola. This species is commonly found on dead or dying twigs of Malus species in Asia, Europe, North America and South Africa (Hayova and Minter 1998a; Adams et al. 2005). It has been associated with dieback of twigs, with symptoms weakened by freeze, wound, and attack from insects and other pathogens (Hayova and Minter 1998a). However, it did not cause significant canker during inoculation tests on apple twigs, contrasting sharply with C. mali and C. pyri (Wang et al. 2007a, 2007b, Wang et al. 2011). C. schulzeri has been found infecting the same apple tree with C. nivea but on adjacent branches in South Africa (Adams et al. 2006). Species identification of C. schulzeri was complicated by its high morphological similarity to C. germanica (Adams et al. 2006) and sharing identical ITS sequences with C. parasitica (Norphanphoun et al. 2017). C. schulzeri has also been found on plants of Castanea, Cerasus, Colutea, Crataegus and Thuja (Fotouhifar et al. 2010; Jiang et al. 2020).
Cytospora parasitica Norphanphoun, Bulgakov & Hyde, 2015
This species was described as a new species on dead and drying branches of M. domestica in Rostov region of Russia (Ariyawansa et al. 2015). It was reported recently that this species caused canker disease on apple in Xinjiang, China (Ma et al. 2018). It is closely related to C. shulzeri with dubious identity. Morphologically, it can be distinguished from C. shulzeri by its single-ostiole conidiomata, contrasting with the multiple-ostiole (2–11) conidiomata of C. shulzeri. There was also some difference in mycelium feature on PDA. However, these two species was indistinguishable in analysis of ITS sequences (Ariyawansa et al. 2015; Norphanphoun et al. 2017). Distinction of two independent clades corresponding to these two species in recent analysis of ITS sequences is a phylogenetic artifact due to inadequate sampling of intraspecific diversity of C. shulzeri (Ma et al. 2018). Indeed, identical ITS sequences were shared by these two species (Fig. 3). Thus, validity of this species remains to be tested based on more genetic markers and isolates.
Cytospora melnikii Norphanphoun, Bulgakov, Wen & Hyde, 2017
This species was recently recognized based on asexual morph on dying branches of M. domestica and Populus nigra from Rostov region of Russia. It is most similar to C. chrysosperma found on Malus but they can be distinguished based on the characteristics of fruiting bodies, conidia size and cultural colonies, and ITS sequences (Norphanphoun et al. 2017). An isolate from Rhus typhina in China (Xinjiang) shares a nearly identical ITS sequence with isolates from Malus and Populus (Fig. 3, Fan et al. 2020). The geographic distribution, host range and pathogenicity of this species remain to be defined.
Cytospora chrysosperma (Pers.) Fr., 1823
The teleomorph state of this species was formerly known as V. sordida. It is a cosmopolitan species and has been known as the causal agent of canker disease on poplars and willows (Kobayashi 1970; Hayova and Minter 1998b; Adams et al. 2006). Its occurrence on Malus has been reported in Iran and China (Fotouhifar et al. 2010; Zang et al. 2012). However, it appears to be genetically uniform with highly similar ITS sequences among isolates from a wide range of hosts (Fig. 3). This species was also present on Armeniaca, Crataegus, Ficus, Fraxinus, Juglans, Ligustrum, Morus, Olea, Persica, Platanus, Populus, Prunus, Robinia, Salix, Tamarix, Thuja, Ulmus and Vitis (Fotouhifar et al. 2010; Zang et al. 2012; Fan et al. 2014; Arzanlou and Narmani 2015; Yang et al. 2015; Lawrence et al. 2017).
Cytospora cincta Sacc., 1884
The sexual form of this species was formerly named as Leucostoma cinctum with variant spelling of L. cincta or Valsa cincta (Adams et al. 2002; Fotouhifar et al. 2010; Mehrabi et al. 2011). This species is a well-known pathogen of stone-fruit tree causing necrosis of twigs and perennial canker, and widely distributed in Asia, Australia, Europe and North America (Hayova and Minter 1998c). Occurrence of C. cincta on Malus hosts has been reported in USA, Iran and China (Proffer and Jones 1989; Surve-Iyer et al. 1995; Adams et al. 2002; Fotouhifar et al. 2010). Although isolates of this species formed a strongly supported monophyletic clade in the phylogenetic tree, their ITS sequences were highly heterogeneous (Fig. 3). Pairwise distances exceeded 2.2% between Malus isolates from USA (AF191170) and Iran (EF447346 and EF447347). In the isozyme analysis, isolates from M. domestica clustered into an independent group distinct from those from Prunus spp. (Surve-Iyer et al. 1995). In addition, cultures of C. cincta from Malus hosts were distinctively colored with reddish brown hues, different from those from Prunus hosts (Proffer and Jones 1989). Thus, it remains to be tested systematically using multiple genes if there are cryptic species within this C. cincta clade. Other hosts of this species include species of Amygdalus, Cerasus, Crataegus, Cydonia, Juglans, Rosa, and Vitis (Fotouhifar et al. 2010).
Cytospora nivea Sacc., 1881
The sexual state of this species was formerly named as Leucostoma niveum or Valsa nivea (Adams et al. 2005). Cytospora nivea is a ubiquitous species on Populus hosts worldwide but appears to be not virulent in some inoculation tests (Adams et al. 2005, 2006). It was first reported occurring on Malus in South Africa and was found to co-infect with C. schulzeri on adjacent branches of the same apple tree (Adams et al. 2006). This species may also occur on Malus in China judging from a highly similar ITS sequence (MG879497) of isolates associated with apple canker in Xinjiang (Fig. 3). However, it still remains to be tested whether this species is a pathogenic agent or an opportunistic endophyte or even saprophyte. Other reported hosts for C. nivea include Elaeagnus, Juglans, and Salix (Fan et al. 2014; Fan et al. 2015; Norphanphoun et al. 2017; Zhao et al. 2018).
Cytospora mali-spectabilis M. Pan & X.L. Fan, 2020
This species was described very recently on branches of Malus spectabilis associated with canker disease from Xinjiang, China (Pan et al. 2020). In the multi-gene phylogenetic analysis, it was most closely related to C. paratranslucens on Populus alba from Rostov region of Russia (Norphanphoun et al. 2017). Its ITS sequence is also highly similar (99.66%) to that of C. nivea and can not be separated from each other in the neighbor-joining tree (Fig. 3).
Cytospora mali-sylvestris Norphanphoun, Bulgakov, T.C. Wen & K.D. Hyde, 2017
This species was introduced after the enforcement of “One fungus = One name”. It was found on dying twigs and branches of Malus sylvestris from Rostov region of Russia. In the combined analysis of ITS and RPB2 sequences, this species was closely related to C. gigaspora isolated from Salix psammophila (Hyde et al. 2017). Its ITS sequence was also highly similar to that of C. melastoma from Malus sp. in USA (Fig. 3), differing only 2 sites in the 538-bp alignment (0.38%).
Cytospora melastoma (Fr.) Sacc., (1882)
This species was recorded occurring on Malus domestica in Michigan (USA) with name epithet Valsella melastoma (Adams et al. 2002). As the genus name Valsella is now treated as synonym of Cytospora (Rossman et al. 2015), this species name was changed into C. melastoma accordingly. Study on this species was very limited with only one ITS sequence available in GenBank (AF191184). This species was thought probably to be a multispored variant of C. nivea (Adams et al. 2006). However, the deep genetic divergence in ITS sequences challenges such an opinion (Fig. 3). It should be noted that its ITS sequence was mislabeled as “Valsella melostoma” in GenBank and in Adams et al. (2006).
Cytospora leucostoma (Pers.) Sacc. 881
The teleomorph state of this species was formerly named as Leucostoma persoonii, Valsa leuscostoma or Valsa persoonii (Adams et al. 2005). It is a common species associated with Rosaceae plants, particularly with Prunus (Adams et al. 2002). Cytospora canker, caused by C. leuscostoma and L. cincta, is one of the most serious diseases of peach worldwide (Chang et al. 1991). C. leuscostoma has been reported from Malus hosts in China, Japan, South Africa and USA (USDA 1960; Adams et al. 2002, 2006; Zang et al. 2012). Genetic heterogeneity was also profound in this species, with divergent ITS sub-lineages being formed but not associated with either geography or host of origins (Fig. 3). This is consistent with earlier isozyme analysis result, in which significant genetic divergences have been documented among isolates from Prunus hosts (Surve-Iyer et al. 1995). A strain CBS367.29 (AF191186, Fig. 3) from Japan labeled as “Valsa mali” by Adams et al. (2002) was in fact part of C. leuscostoma clade (Adams et al. 2005, 2006), but not of C. mali discussed in this paper. Thus detailed investigation is required to test if cryptic divergence or misidentification is involved in this species. Other hosts of C. leuscostoma include plants of Alnus, Betula, Chaenomeles, Cornus, Elaeagnus, Persica, Rosa, Sorbus and Vitis (Adams et al. 2002; Fotouhifar et al. 2010; Kepley et al. 2015; Zhu et al. 2018; Fan et al. 2020).
Concluding remarks and future perspectives
Although Valsa canker poses a serious threat to apple production, our comprehension of this disease has been hampered by confusions on its etiology. A major contributor to such confusions is the changing criteria guiding fungal taxonomy and nomenclature (Guarro et al. 1999). With molecular sequence data emerges as a robust link between the sexual forms and their relevant asexual forms, the process towards “One Fungus = One Name” has been greatly accelerated. However, during the move to “One Fungus = One Name” nomenclature system, great care should be taken to resolve the issue of “One Fungus = Which Name”, so that a stable and meaningful taxonomic system for this economically important fungal group can be agreed upon by all researchers. Resolving this challenge is not only important for scientific communication but also necessary for improved disease management.
Molecular data will play an increasing role in recognition of Cytospora species. However, it is also clear that a single molecular maker (e.g. ITS) is insufficient to provide robust species delineation (Fig. 3). For fungi in general, multilocus data are required for robust delimitation of species boundaries (Dupuis et al. 2012; Wang et al. 2014; Whitehead et al. 2017; Vu et al. 2019). The high diversity in Cytospora suggests an evolutionary history of radiative speciation. The quick succession of the speciation events means that the stochastic nature of the coalescent process in the ancestral species will make different genes have different genealogical histories, i.e. gene trees, due to incomplete lineage sorting of the ancestral polymorphisms (Stewart et al. 2014; Shi and Yang 2018). Thus, it can be expected that gene trees inferred from a single genetic locus will not necessarily be identical to the species tree or the existing ITS-based phylogenetic tree. In addition, interspecies gene flow could further complicate molecular species recognition. New phylogenetic methods that model these biological processes and incorporate gene tree uncertainty (Heled and Drummond 2010; Yang and Rannala 2010; Mirarab et al. 2014; Flouris et al. 2018) should be employed when interrogating multilocus or genome-wide data to delimitate species boundaries and reconstruct phylogeny for this group.
Regional species pool of Cytospora and the major causative species in each region remain to be clarified. It should be systematically evaluated whether occurrence of a given pathogen species on one host poses a threat to other hosts. As different species and strains of Cytospora may differ significantly in their pathogenicity on different hosts (Kepley and Jacobi 2000; Wang et al. 2011), information on pathogenicity of individual strains is pivotal in evaluating their potential threat to apple. Although many Cytospora species are thought to be saprophytes (Fisher 1931), it is unclear whether opportunistic strains would transform into pathogenic ones on new hosts or when they are introduced into new regions or develop an adaptive response to climate change. Thus, monitoring pathogenic modes becomes a necessary preventative step in management of canker disease. Decoding genome sequences (Yin et al. 2015) and identifying genes associated with virulence and pathogenicity of the pathogens (Li et al. 2015; Song et al. 2017; Feng et al. 2017a, 2017b; Wu et al. 2018; Nie et al. 2019; Zhang et al. 2019; Xu et al. 2020), as well as molecules involved in immunological response of hosts (Feng et al. 2017c) will contribute to innovative strategies for management of apple Valsa canker in the future.
Availability of data and materials
Not Applicable.
Abbreviations
- EF1α:
-
Elongation factor-1 alpha
- ICN:
-
International Code of Nomenclature for algae, fungi, and plants
- ITS:
-
Internal transcribed spacer
- rDNA:
-
Ribosomal DNA
References
Abe K, Kotoda N, Kato H, Soejima J. Resistance sources to Valsa canker (Valsa ceratosperma) in a germplasm collection of diverse Malus species. Plant Breed. 2007;126:449–53.
Adams GC, Roux J, Wingfield MJ. Cytospora species (Ascomycota, Diaporthales, Valsaceae): introduced and native pathogens of trees in South Africa. Australas Plant Pathol. 2006;35:521–48.
Adams GC, Surve-Iyer RS, Iezzoni A. Ribosomal DNA sequence divergence and group I introns within the Leucostoma species L. Cinctum, L. persoonii and L. parapersoonii sp. nov., ascomycetes that cause Cytospora canker of fruit trees. Mycologia. 2002;94:947–67.
Adams GC, Wingfield MJ, Common R, Roux J. Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Yalsaceae) from Eucalyptus. Stud Mycol. 2005;52:1–144.
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana WT, Dai DQ, et al. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274.
Arzanlou M, Narmani A. ITS sequence data and morphology differentiate Cytospora chrysosperma associated with trunk disease of grapevine in northern Iran. J Plant Prot Res. 2015;55:117–25.
Ashkan M, Hedjaroude GA. Studies on Cytospora rubescens, a new fungus isolated from apple trees in Iran. Iranian J Plant Pathol. 1993;29:29–30.
Bessho H, Tsuchiya S, Soejima J. Screening methods of apple trees for resistance to Valsa canker. In: Schmidt H, Kellerhals M, editors. Progress in temperate fruit breeding. Dordrecht: Kluwer Academic; 1994. p. 49–52.
Brown-Rytlewski DE, McManus PS. Outbreak of Leucostoma canker caused by Leucostoma cincta on McIntosh apple trees in Wisconsin. Plant Dis. 2000;84:923.
CABI, EPPO. Valsa ceratosperma. In: Distribution Maps of Plant Diseases. Wallingford: CAB International; 2005. Map 954.
Cannon PF, Kirk PM. The philosophy and practicalities of amalgamating anamorph and teleomorph concepts. Stud Mycol. 2000;45:19–25.
Cao KQ, Guo LY, Li BH, Sun GY, Chen HJ. Investigations on the occurrence and control of apple canker in China. J Plant Protec. 2009;35(2):114–6 (in Chinese). http://www.plantprotection.ac.cn/ch/reader/view_abstract.aspx?file_no=200902106.
Chang LS, Iezzoni AF, Adams GC, Ewers FW. Hydraulic conductance in susceptible versus tolerant peach seedlings infected with Leucostoma persoonii. J Am Soc Hortic Sci. 1991;116:831–4.
Chen C, Li M, Shi X, Wang J. Studies on the infection period of Valsa mali Miyabe et Yamada, the causal agent of apple tree canker. Acta Phytopathol Sin. 1987;17(2):65–8 (in Chinese). http://zwblxb.magtech.com.cn/CN/Y1987/V17/I2/65.
Cornille A, Giraud T, Smulders MJM, Roldán-Ruiz I, Gladieux P. The domestication and evolutionary ecology of apples. Trends Genet. 2014;30:57–65.
Crous PW, Groenewald JZ, Slippers B, Wingfield MJ. Global food and fibre security threatened by current inefficiencies in fungal identification. Philos Trans R Soc B. 2016;371:20160024.
Crous PW, Hawksworth DL, Wingfield MJ. Identifying and naming plant-pathogenic fungi: past, present, and future. Annu Rev Phytopathol. 2015;53:247–67.
de Hoog GS, Chaturvedi V, Denning DW, Dyer PS, Frisvad JC, Geiser D, et al. Name changes in medically important fungi and their implications for clinical practice. J Clin Microbiol. 2015;53:1056–62.
Duan N, Bai Y, Sun H, Wang N, Ma Y, Li M, et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat Commun. 2017;8:249.
Dupuis JR, Roe AD, Sperling FAH. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol. 2012;21:4422–36.
EPPO. EPPO global database. In: EPPO global database. Paris: EPPO; 2020.
Fan XL, Bezerra JDP, Tian CM, Crous PW. Cytospora (Diaporthales) in China. Persoonia. 2020;45:1–45.
Fan XL, Hyde KD, Yang Q, Liang YM, Ma R, Tian CM. Cytospora species associated with canker disease of three anti-desertification plants in northwestern China. Phytotaxa. 2015;197:227–44.
Fan XL, Tian CM, Yang Q, Liang YM, You CJ, Zhang YB. Cytospora from Salix in northern China. Mycotaxon. 2014;129:303–15.
Feng H, Xu M, Liu Y, Dong R, Gao X, Huang L. Dicer-like genes are required for H2O2 and KCl stress responses, pathogenicity and small RNA generation in Valsa mali. Front Microbiol. 2017a;8:1166.
Feng H, Xu M, Liu Y, Gao X, Yin Z, Voegele RT, et al. The distinct roles of argonaute protein 2 in the growth, stress responses and pathogenicity of the apple tree canker pathogen. For Pathol. 2017b;47(5):e12354.
Feng H, Xu M, Zheng X, Zhu T, Gao X, Huang L. microRNAs and their targets in apple (Malus domestica cv. “Fuji”) involved in response to infection of pathogen Valsa mali. Front Plant Sci. 2017c;8:2081.
Fisher DF. A Cytospora canker of apple trees. J Agric Res. 1931;43(5):431–8.
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–94.
Flouris T, Jiao X, Rannala B, Yang Z. Species tree inference with BPP using genomic sequences and the multispecies coalescent. Mol Biol Evol. 2018;35(10):2585–93.
Forte AV, Ignatov AN, Ponomarenko VV, Dorokhov DB, Savelyev NI. Phylogeny of Malus (apple tree) species, inferred from the morphological traits and molecular DNA analysis. Russ J Genet. 2002;38:1150–61.
Fotouhifar K-B, Hedjaroude G-A, Leuchtmann A. ITS rDNA phylogeny of Iranian strains of Cytospora and associated teleomorphs. Mycologia. 2010;102:1369–82.
Fuckel L. Enumeratio fungorum Nassoviae, a Leopoldo Fuckel collectorum. Wiesbaden: J Niedner; 1860. https://www.biodiversitylibrary.org/item/22674.
Grove WB. British stem- and leaf-fungi (Coelomycetes), Vol. I Sphaeropsidales. London: Cambridge University Press; 1935.
Guarro J, Gené J, Stchigel AM. Developments in fungal taxonomy. Clin Microbiol Rev. 1999;12:454–500.
Hawksworth DL. Managing and coping with names of pleomorphic fungi in a period of transition. IMA Fungus. 2012;3:15–24.
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, et al. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2:105–12.
Hayova VP, Minter DW. Valsa malicola. IMI descriptions of fungi and bacteria. Set 137, no. 1368. Wallingford: CAB International; 1998a.
Hayova VP, Minter DW. Valsa sordida. IMI descriptions of fungi and bacteria. Set 137, no. 1370. Wallingford: CAB International; 1998b.
Hayova VP, Minter DW. Leucostoma cinctum. IMI descriptions of fungi and bacteria. Set 137, no. 1361. Wallingford: CAB International; 1998c.
Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–80.
Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Chethana KWT, Clericuzio M, et al. Fungal diversity notes 603-708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017;87:1–235.
Ideta A. Handbook of the plant diseases in Japan. 4th ed. Tokyo: Shōkwabō; 1909. p. 295–7.
Janick J, Cummins JN, Brown SK, Hemmat M. Chapter 1 apples. In: Janick J, Moore JN, editors. Fruit breed, volume I: tree and tropical fruits. Hoboken: John Wiley & Sons Inc; 1996. p. 1–77.
Jiang N, Yang Q, Fan XL, Tian CM. Identification of six Cytospora species on Chinese chestnut in China. MycoKeys. 2020;62:1–25.
Ke X, Huang L, Han Q, Gao X, Kang Z. Histological and cytological investigations of the infection and colonization of apple bark by Valsa mali var. mali. Australas Plant Pathol. 2013;42:85–93.
Kepley JB, Jacobi WR. Pathogenicity of Cytospora fungi on six hardwood species. J Arboric. 2000;26:326–33.
Kepley JB, Reeves FB, Jacobi WR, Adams GC. Species associated with Cytospora canker on Populus tremuloides. Mycotaxon. 2015;130(3):783–805.
Kirk PM, Cannon PF, Minter DW, Stalpers JA. Ainsworth & Bisby's dictionary of the Fungi. 10th ed. Wallingford: CAB International; 2008.
Kobayashi T. Taxonomic studies of Japanese Diaportliaccac with special reference to their life-histories. Bull Gov For Exp Stn Meguro. 1970;226:1–242.
Lawrence DP, Travadon R, Pouzoulet J, Rolshausen PE, Wilcox WF, Baumgartner K. Characterization of Cytospora isolates from wood cankers of declining grapevine in North America, with the descriptions of two new Cytospora species. Plant Pathol. 2017;66:713–25.
Lee DH, Lee SW, Choi KH, Kim DA, Uhm JY. Survey on the occurrence of apple diseases in Korea from 1992 to 2000. Plant Pathol J. 2006;22:375–80.
Leonian LH. Studies on the Valsa apple canker in New Mexico. Phytopathology. 1921;11:236–43.
Li Z, Gao X, Du Z, Hu Y, Kang Z, Huang L. Survey of apple Valsa canker in Weibei area of Shaanxi province. Acta Agric Boreali-Occident Sin. 2013;1:029.
Li Z, Yin Z, Fan Y, Xu M, Kang Z, Huang L. Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAXinduced PCD. Front Plant Sci. 2015;6:579.
Lu YJ. Studies on the pathogenic fungus of pear canker disease. Acta Phytopathol Sin. 1992;22(3):197–203 (in Chinese). http://zwblxb.magtech.com.cn/CN/Y1992/V22/I3/197.
Ma R, Liu Y-M, Yin Y-X, Tian C-M. A canker disease of apple caused by Cytospora parasitica recorded in China. For Pathol. 2018;48:e12416.
McNeill J, Barrie FF, Buck WR, Demoulin V, Greuter W, Hawksworth DL, et al., editors. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). [Regnum vegetabile no. 154.]. Königstein: Koeltz Scientific Books; 2012.
Mehrabi M, Mohammadi GE, Fotouhifar KB. Studies on Cytospora canker disease of apple trees in Semiromregion of Iran. J Agr Technol. 2011;7:967–82.
Meng XL, Qi XH, Han ZY, Guo YB, Wang YN, Hu TL, et al. Latent infection of Valsa mali in the seeds, seedlings and twigs of crabapple and apple trees is a potential inoculum source of Valsa canker. Sci Rep. 2019;9:7738.
Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30(17):i541–8.
Miyabe K, Yamada G. Valsa mali, in M. Miura Investigation on apple diseases. Agr Exp Stn Bull (Aomoriken). 1915;15:117–141 (in Japanese).
Nie J, Yin Z, Li Z, Wu Y, Huang L. A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 2019;222:995–1011.
Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:D259–64.
Norphanphoun C, Diolom M, Daranagama DA, Phookamsak R, Wen TC, Bulgakov TS, et al. Revisiting the genus Cytospora and allied species. Mycosphere. 2017;8:51–97.
Ogilvie L. Canker and die-back of apples associated with Valsa ambiens. J Pomol Hortic Sci. 1933;11:205–13.
Old KM, Yuan ZQ, Kobayashi T. A Valsa teleomorph for Cytospora-Eucalypticola. Mycol Res. 1991;95:1253–6.
Paddock W. An apple canker. Science. 1898;8(200):595–6.
Pan M, Zhu H, Bonthond G, Tian C, Fan X. High diversity of Cytospora associated with canker and dieback of Rosaceae in China, with 10 new species described. Front Plant Sci. 2020. https://doi.org/10.3389/fpls.2020.00690.
Peng HX, Wei XY, Xiao YX, Sun Y, Biggs AR, Gleason ML, et al. Management of Valsa canker on apple with adjustments to potassium nutrition. Plant Dis. 2016;100:884–9.
Proffer TJ, Jones AL. A new canker disease of apple caused by Leucostoma cincta and other fungi associated with cankers on apple in Michigan. Plant Dis. 1989;73:508–14.
Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, et al. Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus. 2015;6:145–54.
Rossman AY, Palm-Hernández ME. Systematics of plant pathogenic fungi. Why it matters. Plant Dis. 2008;92:1376–86.
Sawamura K, Harada Y, Yukita K. Apple Valsa canker caused by Valsa ceratosperma (Tode: Fr.) Maire. Bull Fac Agric Hirosaki Univ. 1990;53:10–32.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6.
Shi C-M, Yang Z. Coalescent-based analyses of genomic sequence data provide a robust resolution of phylogenetic relationships among major groups of gibbons. Mol Biol Evol. 2018;35:159–79.
Song N, Dai Q, Zhu B, Wu Y, Xu M, Voegele RT, et al. Gα proteins Gvm2 and Gvm3 regulate vegetative growth, asexual development, and pathogenicity on apple in Valsa mali. PLoS One. 2017;12(3):e0173141.
Spielman LJ. A monograph of Valsa on hardwoods in North-America. Can J Bot. 1985;63:1355–78.
Stevens FL. An apple canker due to Cytospora. Univ Ill Agric Exp Stn Bull. 1919;217:377–9 https://www.ideals.illinois.edu/handle/2142/3151.
Stewart JE, Timmer LW, Lawrence CB, Pryor BM, Peever TL. Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evol Biol. 2014;14:38.
Surve-Iyer RS, Adams GC, Iezzoni AF, Jones AL. Isozyme detection and variation in Leucostoma species from Prunus and Malus. Mycologia. 1995;87:471–82.
Suzaki K. Population structure of Valsa ceratosperma, causal fungus of Valsa canker, in apple and pear orchards. J Gen Plant Pathol. 2008;74:128.
Tai FL. Sylloge fungorum sinicorum, Beijing, China. Beijing: Science Press; 1979. (in Chinese).
Tamura O, Saito I. Histopathological changes of apple bark infected by Valsa ceratosperma (Tode ex Fr.) Maire during dormant and growing periods. Nippon Shokubutsu Byori Gakkaiho. 1982;48:490–8 (in Japanese).
Tanaka T. New Japanese fungi notes and translations: VII. Mycologia. 1919;11:148–54.
Taylor JW. One fungus = one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus. 2011;2:113–20.
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32.
Togashi K. Some studies on a Japanese apple canker and its causal fungus, Valsa mali. J Coll Agric Hokkaido Imp Univ. 1925;12(3):265–324 http://hdl.handle.net/2115/12569.
Turechek WW. Apple diseases and their management. In: Naqvi SAMH, editor. Diseases of fruits and vegetables. Dordrecht: Springer; 2004. p. 1–108.
Turland NJ, Wiersema JH, Barrie FR, Greuter W, Hawksworth DL, Herendeen PS, et al., editors. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. [Regnum Vegetabile no. 159.]. Königstein: Koeltz Botanical Books; 2018.
Turland NJ, Wiersema JH, Munro AM, Deng Y-F, Zhang L. XIX international botanical congress: report of congress action on nomenclature proposals. Taxon. 2017;66:1234–45.
Uhm JY, Sohn HR. Control of apple Valsa canker by localized spraying with neoasozin solution, an arsenic fungicide. Korean J Plant Pathol. 1995;11:9–16.
USDA (United States Departement of Agriculture). Index of plant diseases in the United States. In: Agricultural Handbook; 1960. No.1965. p. 531.
Vasilyeva L, Kim WG. Valsa mali Miyabe et Yamada, the causal fungus of apple tree canker in East Asia. Mycobiology. 2000;28:153–7.
von Höhnel F. Über die Gattung Columnothyrium Bubák. Mitt Bot Inst Tech Hochsch Wien. 1926;3(2):48–50.
Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol. 2019;92:135–54.
Wang L, Zang R, Huang LL, Xie FQ, Gao XN. The investigation of apple tree Valsa canker in Guanzhong region of Shaanxi province. J Northwest Sci-Tech Univ Agric For. 2005;33(Suppl.):98–100 (in Chinese). https://doi.org/10.13207/j.cnki.jnwafu.2005.s1.027.
Wang XL, Kang ZS, Huang LL, Yang P. Pathogen identification of Valsa canker on pear tree: evidences from rDNA-ITS sequences and cultural characteristics. Mycosystema. 2007a;26(4):517–27 (in Chinese). http://manu40.magtech.com.cn/Jwxb/CN/Y2007/V26/I4/517.
Wang XL, Wei JL, Huang LL, Kang ZS. Re-evaluation of pathogens causing Valsa canker on apple in China. Mycologia. 2011;103:317–24.
Wang XL, Zang R, Wang L, Kang ZS, Huang LL. The occurrence of Valsa malicola on apple trees and its pathogenicity. Sci Silvae Sin. 2007b;43(9):23–6 (in Chinese). http://html.rhhz.net/linyekexue/html/20070905.htm.
Wang XL, Zang R, Yin ZY, Kang ZS, Huang LL. Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data. Ecol Evol. 2014;4:1369–80.
Wei JC. Fungal identification manual. Shanghai: Shanghai Science and Technology Press; 1979. (in Chinese).
Whitehead MR, Catullo RA, Ruibal M, Dixon KW, Peakall R, Linde CC. Evaluating multilocus Bayesian species delimitation for discovery of cryptic mycorrhizal diversity. Fungal Ecol. 2017;26:74–84.
Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, et al. Outline of Fungi and fungus-like taxa. Mycosphere. 2020;11:1060–456.
Wu Y, Xu L, Yin Z, Dai Q, Gao X, Feng H, et al. Two members of the velvet family, VmVeA and VmVelB, affect conidiation, virulence and pectinase expression in Valsa mali. Mol Plant Pathol. 2018;19:1639–51.
Xu M, Guo Y, Tian R, Cao C, Guo F, Voegele RT, et al. Adaptive regulation of virulence genes by microRNA-like RNAs in Valsa mali. New Phytol. 2020;227:899–913.
Yang Q, Fang XL, Crous PW, Liang YM, Tian CM. Cytospora from Ulmus pumila in northern China. Mycol Prog. 2015;14:74.
Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci U S A. 2010;107:9264–9.
Yin H, Zhou J-B, Zhang Z-B, Qin N, Ren L, Zhao X-J. Population structure of Valsa species from Malus pumila in Shanxi Province. Mycosystema. 2016;35(12):1493–502 (in Chinese). http://manu40.magtech.com.cn/Jwxb/CN/Y2016/V35/I12/1493.
Yin ZY, Liu HQ, Li ZP, Ke XW, Dou DL, Gao XN, et al. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 2015;208:1202–16.
Zang R, Huang LL, Kang ZS, Wang XL. Biological characteristics and pathogenicity of different isolates of Cytospora spp.isolated from apple trees in Shaanxi province. Acta Phytopathol Sin. 2007;37(4):343–51 (in Chinese). http://zwblxb.magtech.com.cn/CN/Y2007/V37/I4/343.
Zang R, Yin Z, Ke X, Wang X, Li Z, Kang Z, et al. A nested PCR assay for detecting Valsa mali var. mali in different tissues of apple trees. Plant Dis. 2012;96:1645–52.
Zeller SM, Childs L. Perennial canker of apple trees. Oreg Agric Exp Stn Bull. 1925;217.
Zhang M, Xie S, Zhao Y, Meng X, Song L, Feng H, et al. Hce2 domain-containing effectors contribute to the full virulence of Valsa mali in a redundant manner. Mol Plant Pathol. 2019;20:843–56.
Zhang N, Feng H, Fu B, Gao X, Huang L. Incidence of asymptomatic infection of 1-year twigs of apple by Valsa mali in tree provinces of northern China. China Fruits. 2018;4:56–8 (in Chinese).
Zhao SF, Guo KF, He L, Yiming A. First report of Cytospora nivea causing Cytospora canker on walnut (Juglans regia L.) in the Tianshan Mountains Region of Xinjiang, China. Plant Dis. 2018;102(12):2640.
Zhou YX, Cheng LJ, Zhang MX, Zhai LF, Hong N, Wang GP, et al. Sequence analysis and preliminary identification for the pathogen of pear Valsa canker in China. J Fruit Sci. 2013;30(1):140–6 (in Chinese). https://doi.org/10.13925/j.cnki.gsxb.2013.01.007.
Zhu HY, Tian CM, Fan XL. Multigene phylogeny and morphology reveal Cytospora spiraeae sp. nov. (Diaporthales, Ascomycota) in China. Phytotaxa. 2018;338(1):049–62.
Acknowledgments
We would like to thank Dr. Xinli Wei for discussion on fungal nomenclature and Prof. Wen-Ying Zhuang for help with literatures. We are grateful to Prof. Meng Zhang and Prof. Chaoxi Luo as well as three anonymous reviewers for their critical review and constructive suggestions.
Funding
The National Natural Science Foundation of China (Grant No. U1903206) and National Key Research and Development Project of China (Grant No. 2018YFD0201404).
Author information
Authors and Affiliations
Contributions
LH and XW conceived and designed the research. XW, CS and LH collected the data and completed the analyses. XW, CS, MG and LH wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1:
Figure S1. Phylogenetic relationships of Cytospora species illustrate identity of Fan et al.’s (2020) strains. The Bayesian phylogeny shown here reveals that C. pyri is clearly distinct from both C. leucosperma (Valsa ambiens) and C. mali. The phylogeny was constructed using ITS sequences with the best-fit model (TIMef) selected by AIC in Modeltest 3.7. Numbers at the nodes refer to Bayesian posterior probabilities from MrBayes3.2.6 and bootstrap supports from maximum likelihood analysis using IQ-Tree1.6.10.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Shi, CM., Gleason, M.L. et al. Fungal species associated with apple Valsa canker in East Asia. Phytopathol Res 2, 35 (2020). https://doi.org/10.1186/s42483-020-00076-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-020-00076-5