Abstract
Endophytes are promising bio-control agents and rich sources of secondary metabolites known for their biological activities. Two medicinal plants, Pelargonium graveolens and Melia azedarach, and two weeds, Chenopodium album and Malva parviflora, were selected to isolate endophytic fungi. Spore suspension of Alternaria alternata (MG786545), Alternaria solani (MG786543), Curvularia lunata (MF113056), Nigrospora sphaerica (MF113055), and Stemphylium sp. endophytes was tested for their pathogenicity to the third-instar larvae of the cotton leaf worm, Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae). Two techniques were used: dipping in spore suspension and residual film assay. Secondary metabolites were extracted, and ethyl acetate extracts of A. alternata (MG786545), C. lunata (MF113056), and N. sphaerica (MF113055) were tested for their insecticidal activity by both techniques. C. lunata (MF113056) showed the most potent pathogenicity causing 60% mortality after 7 days by dipping bioassay, while A. solani (MG7865453) and A. alternata (MG786545) caused 40 and 33.3% mortality, respectively. All tested endophytes showed no effect against S. littoralis by residual film method. N. sphaerica ethyl acetate extract activated larval feeding on castor leaves causing 25% mortality by residual film at 2000 mg/l.
Similar content being viewed by others
Background
The extensive use of conventional pesticides has led to serious ecological problems including hazards to human health and animals, resistance of target pests, and environmental pollution. Bio-pesticides are natural pesticides derived from animals, plants, microorganisms, and certain minerals. They are eco-friendly tools of managing agriculture pests, less toxic, and specific on target organisms (Gupta and Dikshit 2010). Microbial pesticides’ active ingredients are microorganisms such as fungus, bacterium, virus, protozoan, or algae that have a broad spectrum of activity, but every organism is relatively specific to its target pest (Lord 2001). Beauveria bassiana is one of the most common fungal examples of successful entomopathogenic used as a bio-control of several insect pests and has been registered by the US Environmental Protection Agency (EPA) as a bio-control agent for a wide range of insect pests (Salma et al. 2011).
Endophytes are microorganisms that grow within plants without causing any obvious symptoms of infection or disease. Some endophytes accelerate the growth of their host plants and increase nutrient uptake (Hashem et al. 2016). Other endophytes protect their hosts by producing secondary metabolites which have the potential to kill or suppress the attack by pathogenic microorganisms or herbivores (Carroll 1988 and Schulz et al. 2002). So, endophytes can be used as one of the biological control agents in sustainable crop production (Rybakova et al. 2016 and Egamberdieva et al. 2017).
The Egyptian cotton leafworm, Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae), is a serious lepidopteron pest in subtropical and tropical regions. It has a wide range of host plants belonging to 44 different families including grasses, legumes, and cruciferous and deciduous fruit trees (Abdel-Megeed and Iss-Hak 1975).
The aims of this work were to isolate endophytic fungi from Egyptian weeds and medicinal plants, to extract their secondary metabolites, and to evaluate their potential against S. littoralis larvae.
Materials and methods
Isolation of endophytic fungi
Root samples of Malva parviflora and leaf samples of Chenopodium album and Pelargonium graveolens were collected from Abees farm, and leaf samples of Melia azedarach were collected from the garden of the Faculty of Agriculture, Alexandria, Egypt. The samples were washed with tap and distilled water, air dried, surface sterilized by 70% ethanol for 1 min then by 3.0% sodium hypochlorite (NaOCl) for 3 min, and rinsed with sterile distilled water for 1 min three times. The surface-sterilized samples were cut into 5 mm2 surface using a sterile blade and placed in Petri dishes with potato dextrose agar media containing 50 mg/l of ampicillin (Kharwar et al. 2012). Each plate contained four segments of plant pieces with four replicates for each plant. The dishes were incubated at 25 ± 2 °C and observed daily to record the emergence of endophytes and pre-cultured on potato dextrose agar (PDA) media. For testing the effectiveness of surface-sterilized plant samples, 1 ml of the last rinsing water was placed on the PDA media and incubated at 25 ± 2 °C.
Identification of fungal isolates
Morphological observations were carried out, using both macroscopic and microscopic characters, for endophytic fungus culture and spore shapes and types, using the standard manual of Barnett and Hunter (1998). The isolated endophytes were subjected to DNA extraction, using the Qiagen DNA extraction kit (Qiagen, Germany). According to Hafez and Elbestawy (2009), endophytic fungi were identified by amplification of ITS1-5.8SrRNA-ITS4 gene using the universal primers: ITS1 (forward primer: 5-TCC GTA GGT GAA CCT GCG G-3) and ITS4 (reverse primer: 5-TCC TCC GCT TAT TGA TAT G-3). PCR reaction mixture (25 μl) consisted of 12.5 μl PCR Master Mix (2X) Promega Corporation, 1 μl (50 pmol) of each oligonucleotide primer, 1 μl DNA template, and 9.5 μl nuclease-free water. The reaction cycles consisted of initial denaturation (95 °C, 2 min), 35 cycles of denaturation (95 °C, 31 min), annealing (55 °C, 1 min), extension (72 °C, 1 min), and final extension (72 °C, 10 min). The PCR products were analyzed using agarose gel electrophoresis with ethidium bromide (EtBr) staining. The products were purified using the purification kit (Qiagen) and sequenced by DNA sequencer by LGC group (Berlin, Germany). After preprocessing, the resulting DNA sequence was identified using the BLASTN tool of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) and nucleotide collection (nr/nt) database. To create the phylogenetic tree of the endophytic fungi, advanced version of the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 was used (Tamura et al. 2013).
Pathogenicity of endophytic fungi spore suspension
By dipping technique
The endophytic fungi grown in PDA plates with high spore production levels (Alternaria alternata (MG786545), A. solani (MG786543), Curvularia lunata (MF113056), Nigrospora sphaerica (MF113055), and Stemphylium sp.) were flooded by sterile distilled water containing 0.05% Triton X-100, scrapped with surface-sterilized spatula, and filtered through cheesecloth to remove mycelial debris. Spore suspensions were prepared, and their concentrations were adjusted to 108 spores ml−1 by a hemocytometer then tested against third-instar larvae of S. littorals. Ten larvae were dipped in spore suspensions for 5 s and left to dry on a filter paper. Two control treatments were conducted: with sterilized distilled water and with aqueous solution of 0.05% Triton X-100. The treated larvae were transferred to glass bottles containing fresh castor leaves, with four replicates for each treatment (Baskar et al. 2012). The experiment was checked daily for 2 weeks to keep clean, the castor leaves were changed, the dead larvae were counted, and the mortality percent was calculated. The dead larvae were surface sterilized and placed on the Petri dishes containing PDA to confirm if the mortality was due to the fungi pathogenicity.
By residual film
The same concentration of spore suspensions used for the previous bioassay was used for residual film bioassay. Castor leaves were submerged in spore suspensions for 20 s, left to dry, and transferred to glass bottles with ten starved (4–5 h) third-instar larvae of S. littoralis. Castor leaves treated with sterilized distilled water and with sterilized distilled water containing 0.05% Triton X-100 served as the control treatments with four replicates for all treatments. The results were taken as described above.
Extraction of endophytes secondary metabolites
Based on the results of the previous pathogenicity bioassay, three endophytic isolates from different genera were chosen for fermentation and extraction of their secondary metabolites. Five disks of each endophytic fungus were inoculated in 1 l Erlenmeyer flask containing 300 ml potato dextrose broth (3 l PDB) and incubated at 28 °C for 2 weeks in a shaker incubator at 120 rpm. The culture broth was centrifuged for 10 min at 15000×g, and the supernatant was extracted by the solvent mixture (ethyl acetate to methanol to water, 8:4:1) in an ultrasonic bath at 40 °C for 20 min. The solvent layers were separated by a separatory funnel, and the aqueous layer was extracted twice by ethyl acetate with sonication for 20 min. The ethyl acetate layer was collected and evaporated under vacuum to dry and kept at 4 °C for bioassay.
Pathogenicity of endophyte secondary metabolites
Ethyl acetate extracts were tested against third-instar larvae of S. littoralis by dipping and residual film techniques (Pampapathy et al. 2011). Concentrations of ethyl acetate crude extracts (250, 500, 1000, 2000 mg/l) were prepared in a sterile distilled water containing 0.05% Triton X-100. Ten larvae of S. littoralis were dipped in each extract concentrations for 5 s, left on filter paper for 10 s to dry, and transferred to the Petri dishes containing castor leaves, with four replicates.
The same concentrations of ethyl acetate extracts were used for residual film bioassay. Castor leaves were dipped in each extract concentrations for 20 s, left to dry, and transferred to the Petri dishes containing ten starved (4–5 h) larvae of S. littoralis with four replicates. Control treatments were conducted using castor leaves treated with sterilized distilled water and with sterilized distilled water containing 0.05% Triton X-100. Mortality rate was observed daily for 1 week, and mortality percent was calculated.
Statistical analysis
Percentage of S. littoralis mortality was subjected to one-way analysis of variance followed by Student-Newman-Keuls test (Cohort Software Inc. 1985) to determine significant differences between mean values at the probability level of 0.05.
Results and discussion
Identification of the endophytic isolates
Fungal identification by morphological characters resulted in the identification of ten endophytic fungi. Eight of them were identified by DNA sequences of the purified PCR product. Five endophytic fungi were isolated from C. album weed: F. chlamydosporum saad3 MG786540, A. alternata saad5 MG786542, A. alternata saad8 MG786545, F. oxysporum saad4 MG786541, and Phoma sp. Two were isolated from M. parviflora weed: F. equiseti saad7 MG786544 and Stemphylium sp. Two were isolated from the medicinal plant M. azedarach: C. lunata saad2 MF113056 and N. sphaerica saad1 MF113055. The last one was isolated from the aromatic plant P. graveolens: A. solani saad6 MG786543 (Table 1). Figures 1 and 2 illustrate the gel electrophoresis and phylogenetic tree of the isolated endophytic fungi, respectively.
Pathogenicity of endophytic fungi and their metabolites
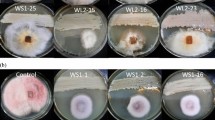
The results of spore suspension bioassays indicated that the mortality of S. littoralis larvae differed according to the fungus species and application method. The endophytic isolate pathogenicity was confirmed after the fungi mycelium emerged from the dead larvae (Fig. 3). In dipping method, C. lunata (MF113056) spore suspension showed the most virulence with 60% mortality rate after 7 days. A. solani (MG7865453) and A. alternata (MG786545) caused 40% and 33.3% mortality rate, respectively, but there was no significant difference between them. In contrast, N. sphaerica showed no virulence by both techniques (Fig. 4). The mortality did not increase after 1 week until the end of the experiment for all used endophytes. All the tested endophytes showed very weak or no pathogenicity by residual film method, and there was no difference between endophyte treatments and control treatments regarding larval mortality. The ethyl acetate extracts showed no effects in S. littoralis mortality by dipping method and weak effects by residual film method. Only ethyl acetate crude extract of N. sphaerica activated the feeding on castor leaves and caused 25% mortality rate by the residual film method at 2000 mg/l (Fig. 5).
Many researchers have reported the endophytic fungi as pathogenic to insects and can protect their host plants from herbivores by producing secondary metabolites (Schardl 2001; Baskar et al. 2012; and Bensaci et al. 2015). Senthilkumar et al. (2014) found that the methanol extracts (1000 ppm) of Aspergillus flavus and Nigrospora sphaerica isolated from Tectona grandis caused 65% and 62% mortality of Hyblea purea, 59% and 56% of Eligma narcissus, and 46% and 42% of Atteva fabriciella, respectively. Han et al. (2013) stated that ethyl acetate extracts of both 16 strains mycelium and 9 strains fermentation broth showed a larvicidal activity against the third-instar larvae of Culex pipiens pallens mosquitos. These active strains were focused in the genera Aspergillus, Penicillium, Phoma, and Pesalotiopsis. However, the obtained results showed that C. lunata (MF113056) and A. alternata (MG786545) had the potential to infect and cause the death of S. littoralis larvae by direct contact not by consuming fungal spores or by their metabolites. This finding is in agreement with the entomopathogenic fungal mechanism of action, as it invades directly through the cuticle, mechanically or by enzymatic production without a need to be consumed by their host to cause infection (Ownley et al. 2008, 2010, and Vega 2008, Vega et al. 2012).
In conclusion, the obtained finding showed the potential of C. lunata (MF113056), A. solani (MG7865453), and A. alternata (MG786545) to act as entomopathogenic fungi and cause 60%, 40%, and 33.3% death of S. littoralis larvae, respectively. Therefore, endophytic fungi represent a group of organisms with a potential for applications as one of the bio-control agents against insect pests.
References
Abdel-Megeed MI, Iss-Hak RR (1975) Field observations on the vertical distribution of the cotton leafworm, Spodoptera littoralis (Boisd.) on cotton plants. Zeitschr f Angew Ent 78:59–62
Barnett HL, Hunter BB (1998) Illustrated genera of imperfect fungi, 4th edn. APS Press, St. Paul, p. 218
Baskar K, Raj GA, Mohan PM, Lingathurai S, Ambrose T, Muthu C (2012) Larvicidal and growth inhibitory activities of entomopathogenic fungus, Beauveria bassiana against Asian army worm, Spodoptera litura Fab. (Lepidoptera: Noctuidae). J Entomol 9:155–162
Bensaci OA, Daoud H, Lombarkia N, Rouabah K (2015) Formulation of the endophytic fungus Cladosporium oxysporum Berk. & M.A. Curtis, isolated from Euphorbia bupleuroides subsp. luteola, as a new biocontrol tool against the black bean aphid (Aphis fabae Scop.). J Plant Protect Res 55:80–87
Carroll GC (1988) Fungal endophytes in stems and leaves: from latent pathogens to mutualistic symbiont. Ecol 69:2–9
Egamberdieva D, Wirth SJ, Shurigin VV, Hashem A, Abd Allah EF (2017) Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under salt stress. Front Microbiol 8:1887
Gupta S, Dikshit AK (2010) Biopesticides: an ecofriendly approach for pest control. J Biopest 3:186–188
Hafez E, Elbestawy E (2009) Molecular characterization of soil microorganisms: effect of industrial pollution on distribution and biodiversity. W.J Microbiol Biotechnol 25:215–224
Han LR, Wang ZH, Zhang HJ, Xue LS, Feng JT, Zhang X (2013) Isolation of endophytic fungi from Tripterygium wilfordii and their insecticidal activities. Afr J Microbiol Res 7:771–776
Hashem A, Abd_Allah EF, Alqarawi A, Al-Huqail AA, Wirth S, Egamberdieva D (2016) The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acaciagerrardii under salt stress. Front Plant Sci 7:1089
Kharwar RN, Maurya AL, Verma VC, Kumar A, Gond SK, Mishra A (2012) Diversity and antimicrobial activity of endophytic fungal community isolated from medicinal plant Cinnamomum camphora. Proc Natl Acad Sci, India, Section B: Biol Sci 82:557–565
Lord JC (2001) Desiccant dusts synergize the effect of Beauveria bassiana (Hyphomycetes: Moniliales) on stored-grain beetles. J Econom Entomol 94:367–372
Ownley BH, Griffin MR, Klingeman WE, Gwinn KD, Moulton JK, Pereira RM (2008) Beauveria bassiana: endophytic colonization and plant disease control. J Invert Pathol 98:267–270
Ownley BH, Gwinn KD, Vega FE (2010) Endophytic fungal entomopathogens with activity against plant pathogens: ecology and evolution. Biocont 55:113–128
Pampapathy G, McGee PA, Sword G (2011) Endophytic Lecanicillium lecanii and Beauveria bassiana reduce the survival and fecundity of Aphis gossypii following contact with conidia and secondary metabolites. Crop Protect 30:349–353
Rybakova D, Cernava T, Köberl M, Liebminger S, Etemadi M, Berg G (2016) Endophytes-assisted biocontrol: novel insights in ecology and the mode of action of Paenibacillus. J. Plant Soil 405:125–140
Salma M, Ratul CR, Jogen CK (2011) A review on the use of biopesticides in insect pest management. Int J Sci Adv Tech 1:169–178
Schardl CL (2001) Epichloe festucae and related mutualistic symbionts of grasses. Fung Gen Biol 33:69–82
Schulz B, Boyle C, Draeger S, Römmert A-K, Krohn K (2002) Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol Res 106:996–1004
Senthilkumar N, Murugesan S, Babu DS (2014) Metabolite profiling of the extracts of endophytic fungi of entomopathogenic significance, Aspergillus flavus and Nigrospora sphaerica isolated from tropical tree species of India, Tectona grandis L. J Agric Life Sci 1:108–114
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Vega FE (2008) Insect pathology and fungal endophytes. J Invert Pathol 98:277–279
Vega FE, Meyling NV, Luangsa-ard JJ, Blackwell M (2012) Fungal entomopathogens. In: Vega FE, Kaya HK (eds) Insect pathology, 2nd edn. Academic Press, San Diego, pp 171–220
Acknowledgements
My great appreciations are to Prof. Samir A.M. Abdelgaleil, Dr. Yousf Moustafa and Dr. Doaa Komeil for their help and assistance whenever I needed during this work.
Funding
Faculty of agriculture, Alexandria University.
Availability of data and materials
Not applicable in this section.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design of the experiment, acquisition of data, analysis and interpretation of data, and drafting and revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable in this section.
Consent for publication
Not applicable in this section.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Saad, M.M.G., Ghareeb, R.Y. & Saeed, A.A. The potential of endophytic fungi as bio-control agents against the cotton leafworm, Spodoptera littoralis (Boisd.) (Lepidoptera: Noctuidae). Egypt J Biol Pest Control 29, 7 (2019). https://doi.org/10.1186/s41938-019-0108-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-019-0108-x