Abstract
Background
Non-small cell lung cancer (NSCLC) is one of the leading causes of death in the world. NSCLC diagnosed at an early stage can be highly curable with a positive prognosis, but biomarker limitations make it difficult to diagnose lung cancer at an early stage. To identify biomarkers for lung cancer development, we previously focused on the oncogenic roles of transcription factor TFAP2C in lung cancers and revealed the molecular mechanism of several oncogenes in lung tumorigenesis based on TFAP2C-related microarray analysis.
Results
In this study, we analyzed microarray data to identify tumor suppressor genes and nine genes downregulated by TFAP2C were screened. Among the nine genes, we focused on growth arrest and DNA-damage-inducible beta (GADD45B) and phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1) as representative TFAP2C-regulated tumor suppressor genes. It was observed that overexpressed TFAP2C resulted in inhibition of GADD45B and PMAIP1 expressions at both the mRNA and protein levels in NSCLC cells. In addition, downregulation of GADD45B and PMAIP1 by TFAP2C promoted cell proliferation and cell motility, which are closely associated with NSCLC tumorigenesis.
Conclusion
This study indicates that GADD45B and PMAIP1 could be promising tumor suppressors for NSCLC and might be useful as prognostic markers for use in NSCLC therapy.
Similar content being viewed by others
Background
Lung cancer is one of the most common cancers and is the leading cause of cancer-related deaths worldwide. Lung cancer is divided into small cell lung cancer and non-small cell lung cancer (NSCLC) with NSCLC responsible for almost 80% of lung cancer-related deaths [1, 2]. Despite improvements in chemotherapy and molecular-targeted therapy over recent decades, the 5-year survival rate of NSCLC patients is only 14% [2, 3]. Patients with NSCLC can have a greatly improved prognosis and survival rate when they are diagnosed at an early stage. However, early diagnosis of NSCLC is difficult due to biomarker limitations. Discovery of tumor-specific factors that can influence the diagnosis of NSCLC is critical. Thus, a study to identify novel molecular targets of NSCLC can contribute to improvements in the early diagnosis and therapy of NSCLC.
Transcription factor activating enhancer-binding protein 2C (TFAP2C) is a member of the AP-2 family which has five isoforms. TFAP2C was first shown to play a role in embryonic development [4]. Subsequently, accumulated data have provided several clues that TFAP2C is also highly involved in human solid cancers. TFAP2C is responsible for angiogenesis via an increase in the expression of the extracellular matrix 1 gene in melanoma [5]. In luminal breast cancer, TFAP2C increases epidermal growth factor receptor expression, resulting in cancer cell proliferation and tumor growth [6]. To elucidate a role of TFAP2C in NSCLC, we previously reported that TFAP2C mediates inhibition of A-kinase anchor protein 12 and activation of cell division protein kinase 6 through upregulation of miR-183 and downregulation of miR-33a, leading to cell cycle progression and NSCLC malignancy [7]. TFAP2C can also increase the expression level of transforming growth factor beta receptor I, consequently promoting NSCLC cell proliferation [8]. Recently, it was reported that TFAP2C participates in the enhancement of cancer stemness and chemoresistance in colorectal cancer [9]. These studies have shown that TFAP2C is significantly involved in various cancers; an involvement that may depend on environmental conditions and tumor type. Although these previous studies have indicated the importance of TFAP2C in cancer progression, its roles in NSCLC remain to be investigated.
The present study was established to identify novel tumor suppressive genes that are downregulated during TFAP2C-mediated NSCLC tumorigenesis. Through an analysis of a TFAP2C-related microarray dataset, we identified nine candidate genes functioning as tumor suppressors. In particular, growth arrest and DNA-damage-inducible beta (GADD45B) and phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1) were downregulated by TFAP2C-overexpressing NSCLC cells, which contributed to NSCLC cell proliferation and motility. Our analysis has identified putative target genes for TFAP2C-mediated NSCLC tumorigenesis, and the results suggest that those genes might be used to provide prognostic targets in the treatment of NSCLC.
Methods
Antibodies and reagents
Antibodies specific for TFAP2C (sc-12762), tubulin (sc-23948), GADD45B (sc-377311) and PMAIP1 (sc-515840) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cell culture media (DMEM and RPMI-1640), fetal bovine serum (FBS), penicillin and streptomycin were acquired from Gibco (Grand Island, NY, USA). Small interfering RNA (siRNA) specific for TFAP2C (sc-29696), GADD45B (sc-37416), and PMAIP1 (sc-37305) with control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology.
Cell lines and cell culture
The human normal lung cell line, WI-26 VA4, and four human NSCLC cell lines, A549, NCI-H292, NCI-H358, and NCI-H460, were acquired from the American Type Culture Collection (ATCC, Manassas, VA), authenticated, and then maintained in early passages for no more than 6 months after receipt from ATCC. The WI-26 VA4, A549, NCI-H460, NCI-H292, and NCI-H358 cells were grown in DMEM or RPMI-1640 supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a 5% CO2/95% air atmosphere.
Transient transfection and real-time reverse transcriptase polymerase chain reaction (qRT-PCR)
A plasmid containing a full-length TFAP2C construct (pcDNA3.1-TFAP2C) and a control plasmid (pcDNA3.1 empty vector) were kindly provided by Ronald J. Weigel (University of Iowa, Iowa City, IA, USA). Cells were plated in 60 mm dishes before undergoing transfection. The cells were transiently transfected with pcDNA3.1-TFAP2C by using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) and with si-TFAP2C, si-GADD45B, and/or si-PMAIP1 by using Lipofectamine 3000 (Invitrogen). After 48 h of transfection, the cells were collected and used for in vitro functional analyses. To analyze the expression levels of mRNAs [10], total RNA was isolated from cells using TRIzol (Invitrogen). To obtain cDNA, the isolated RNA was converted by using an MMLV Reverse Transcriptase system (Bioneer, Daejeon, Republic of Korea) according to the manufacturer’s protocol. The cycle parameters for the reverse transcription (RT) reaction were 25 °C for 5 min, 37 °C for 60 min, 95 °C for 5 min, and then held at 4 °C until used. Each RT product was used as a template for real-time quantitative PCR (qRT-PCR) with specific primers (Table 1). Aliquots of a master mix containing all reaction components and the primers were dispensed into a real time PCR plate (Applied Biosystems, Foster City, CA, USA). All PCR reagents were obtained from a SYBR Green core reagent kit (Applied Biosystems). Gene mRNA levels were measured in triplicate in the reaction plate. Real-time qRT-PCR was performed using an Applied Biosystems-7900 HT qRT-PCR instrument. PCR was performed over 40 cycles of 15 s at 95 °C and 1 min at 60 °C, after which samples were subjected to thermal denaturation. Gene expression levels versus ACTN mRNA were determined using the 2−ΔΔCT method [11]. To simplify data presentation, relative expression values were multiplied by 102.
Western blot analysis
After experimental treatments, Western blotting was performed as previously described [12]. Whole-cell lysates and tissue lysates were obtained by using an EzRIPA Lysis kit (ATTO, Tokyo, Japan; 20 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholic acid, as well as protease inhibitor and phosphatase inhibitor cocktails). Protein concentrations in lysates were determined by using a BioRad protein assay kit (BioRad Laboratories, Hercules, CA, USA). SDS-PAGE was performed with the protein samples, after which the proteins were transferred to a nitrocellulose membrane. For blocking, 5% bovine serum albumin in TBST (10 mM Tris, 100 mM NaCl, and 0.1% Tween 20) was used for 1 h at room temperature. Membranes were then incubated at 4 °C overnight with specific primary antibodies and subsequently probed with peroxidase-conjugated secondary antibodies (Santa Cruz) for 1 h at room temperature. Blots were visualized by using an ECL detection system (Abfrontier, Seoul, Republic of Korea). Data acquisition and densitometric analysis were performed using an iBright CL1000 imaging system (Thermo Fisher Scientific, Waltham, MA, USA).
Cell viability assay
For cell viability assays, cells were cultured in 35-mm dishes at a density of 5 × 104 cells. After 24 h, they were treated with TFAP2C overexpression and/or siRNA transfection. Cells were detached with 10% trypsin–EDTA and washed with PBS at the indicated times. The cells were resuspended in PBS and diluted 1:1 in trypan blue solution (Gibco). Cell viability was measured by determining the number of viable cells that excluded trypan blue solution.
Colony-forming assay
For colony-forming assays [13], cells were plated at a density of 1000 cells per 35 mm dish. After 24 h, the cells were treated with the indicated gene and/or siRNA and then incubated at 37 °C in a 5% CO2/95% air atmosphere for 14 days. Subsequently, cells were fixed with 10% methanol, 10% acetic acid and stained with 1% crystal violet (Sigma, St. Louis, MO, USA). Colonies containing more than 50 cells were identified by using ImageJ software [14] and scored as survivors.
Apoptosis assay
A Caspase-Glo 3/7 assay kit (Promega, Madison, WI) was used to detect apoptosis [15]. Briefly, treated cells (104 cell per mL) in 100 μL of culture medium were transferred to a 96-well microplate, 100 μL of Caspase-Glo 3/7 reagent, which contained caspase 3/7 substrates, was added to each well. Well contents were gently mixed at 300 to 500 rpm for 30 s, and incubated at room temperature for 1 h. Sample luminescence was measured using a Glomax multidetection system (Promega).
Wound-healing assay
For wound-healing assays [16], cells were cultured to 70% confluency in DMEM or RPMI-1640 medium with 1% FBS. The cell monolayers were then scratched with a 200 μL pipette tip. The cells were further incubated with fresh medium with or without treatment for 24 h or 48 h. Photomicrographs were taken at × 100 magnification with an Olympus CKX53 inverted microscope (Olympus Optical, Tokyo, Japan).
Transwell cell migration assay
For cell migration assays [16, 17], cells (1 × 104 in serum-free DMEM or RPMI-1640 medium) subjected to the desired treatments for 72 h were seeded into the upper chambers of a 24-well Transwell chamber (Corning, Corning, NY) fitted with a 5 μm pore-size insert. The lower chamber was filled with DMEM or RPMI-1640 medium containing 2% FBS. After 6 h, the upper membrane surface was wiped with a cotton swab to remove cells that had not migrated into the lower chamber. Cells that had migrated to the lower membrane surface were fixed with 4% paraformaldehyde, stained with hematoxylin, and counted. Migration indices were calculated and normalized to the number of control cells that had migrated. Results are expressed as fold increases in migration compared to control groups as determined by relative numbers of cells in a randomly selected field in experiments conducted in triplicate.
Statistical analysis
All numeric data are presented as mean ± standard deviation (SD) values from at least three independent experiments and sample sizes were calculated to allow significance to be determined. Experimental results were analyzed by one-way ANOVA for ranked data followed by Tukey’s honestly significant difference test and by two-way ANOVA for ranked data followed by Bonferroni’s post-test. Prism 5 software (GraphPad Software, San Diego, CA, USA) was used to conduct all statistical analyses, and statistical significance was accepted for a p value < 0.05.
Results
Screening candidates for tumor suppressor genes regulated by TFAP2C in lung tumorigenesis
Based on the role of TFAP2C in lung tumorigenesis identified in our previous work, we focused on other genes transcriptionally regulated by TFAP2C [7, 8]. Microarray data were analyzed for TFAP2C-affected genes in NSCLC cells by using the Gene Expression Omnibus database (GEO Series accession number GSE79228). As shown in Fig. 1, among the many genes upregulated or downregulated by TFAP2C knockdown (KD-TFAP2C, > 1.5-fold), we were interested in potential tumor suppressor genes. Of the 1152 genes upregulated by KD-TFAP2C, we excluded pseudogenes and nonfunctional genes. Of the 58 genes screened by analysis of gene ontology functional annotation based on hallmarks of cancer including apoptosis, anti-proliferation, and cell death, nine genes (GADD45B, PMAIP1, XAF1, CYR61, IL24, ATF3, DLC1, RHOB, and TNFAIP3) have been reported to act as tumor suppressive genes in several cancer types (Table 2).
Verification of downregulation of selected tumor suppressor genes in NSCLC
Next, to verify our results for the screening of nine genes as tumor suppressors for NSCLC, the genes were further investigated using Oncomine (http://www.oncomine.org), an online database for expression analysis of lung cancer tissues [18]. The database results indicated that GADD45B was downregulated by − 8.080-fold in lung adenocarcinomas [19], PMAIP1 was downregulated by − 1.067-fold in lung adenocarcinomas [20], XAF1 was downregulated by − 1.441-fold in large cell lung carcinomas [21], CYR61 was downregulated by − 2.381-fold in lung adenocarcinomas [22], IL24 was downregulated by − 1.972-fold in lung adenocarcinomas [19], ATF3 was downregulated by − 2.589-fold in lung adenocarcinomas [23], DLC1 was downregulated by − 2.541-fold in lung adenocarcinomas [24], RHOB was downregulated by − 1.950-fold in lung adenocarcinomas [23], and TNFAIP3 was downregulated by − 1.901-fold in lung adenocarcinomas [22], compared to normal lung counterparts, respectively (each p < 0.05, Fig. 2). These results showed that the nine genes were expressed at low levels in NSCLC tissues, indicating that they might provide clues for using the nine genes as potential prognostic markers for NSCLC tumor suppressors. Based on the results that GADD45B expression level was the lowest (− 8.080-fold change) in Oncomine data and PMAIP1 expression was highest (1.741-fold, data not shown) in KD-TFAP2C mediated microarray data, we selected GADD45B and PMAIP1 to represent the nine candidate genes in further study.
The expression patterns of nine selected genes in NSCLC based on analysis of the Oncomine database. a The Bhattacharjee dataset showed GADD45B downregulation in lung adenocarcinoma compared with the normal lung counterparts (fold change = –8.080, p = 5.52E−5). b The Weiss dataset showed PMAIP1 downregulation in lung adenocarcinoma (fold change = − 1.067, p = 4.15E−6). c The Hou dataset showed XAF1 downregulation in lung adenocarcinoma (fold change = − 1.441, p = 9.90E−5). d The Selamat dataset showed CYR61 downregulation in lung adenocarcinoma (fold change = − 2.381, p = 5.54E−10. e The Bhattacharjee dataset showed IL24 downregulation in lung adenocarcinoma (fold change = − 1.972, p = 0.011). f The Landi dataset showed ATF3 downregulation in lung adenocarcinoma (fold change = − 2.589, p = 1.70E−8). g The Okayama dataset showed DLC1 downregulation in lung adenocarcinoma (fold change = − 2.541, p = 3.04E−13). h The Landi dataset showed RHOB downregulation in lung adenocarcinoma (fold change = − 1.950, p = 3.03E−11). i The Selamat dataset showed TNFAIP3 downregulation in lung adenocarcinoma (fold change = − 1.901, p = 4.03E−9)
Downregulation of GADD45B and PMAIP1 expression by TFAP2C in NSCLC cells
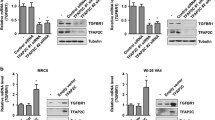
TFAP2C is primarily upregulated in NSCLC cells and tissues but downregulated in normal lung counterparts [7]. To confirm the microarray results, we analyzed GADD45B and PMAIP1 levels in TFAP2C-overexpressing normal lung cells (WI-26 VA4 cells) and TFAP2C siRNA-treated NSCLC cells (NCI-H292, NCI-H358, NCI-H460, and A549 cells) by using real time quantitative reverse transcription PCR (qRT-PCR). The mRNA levels of GADD45B and PMAIP1 were significantly decreased by TFAP2C overexpression in WI-26 VA4 cells (Fig. 3a) but were increased by treatment of TFAP2C siRNA in the four human NSCLC cell lines (Fig. 3b–e). Similarly, protein levels of GADD45B and PMAIP1 were downregulated in TFAP2C-overexpressing WI-26 VA4 cells (Fig. 3f) but were upregulated in TFAP2C siRNA-transfected NSCLC cells (Fig. 3g). These results suggest that GADD45B and PMAIP1 expressions were inhibited by TFAP2C and negatively correlated with the expression of TFAP2C.
TFAP2C induces GADD45B and PMAIP1 downregulation in NSCLC cells. a The mRNA expression levels of GADD45B and PMAIP1 in TFAP2C-overexpressing WI-26 VA4 cells were examined by qRT-PCR. *p < 0.05 compared to non-treated cells or cells treated with control vector. b–e The mRNA expression levels of GADD45B and PMAIP1 in TFAP2C siRNA-treated NCI-H292 (b), NCI-H358 (c), NCI-H460 (d), and A549 (e) cells were examined by qRT-PCR. *p < 0.05 compared to non-treated cells or cells treated with control siRNA. f The protein expression levels of GADD45B and PMAIP1 in TFAP2C-overexpressing WI-26 VA4 cells were examined by Western blotting. g The protein expression levels of GADD45B and PMAIP1 in TFAP2C siRNA-treated NCI-H292, NCI-H358, NCI-H460, and A549 cells were examined by Western blotting
Increased cell proliferation in NSCLC cells by downregulation of GADD45B and PMAIP1 expression
It has been reported that GADD45B and PMAIP1 have significant roles in anti-proliferation and apoptosis, suggesting that they may function as tumor suppressors [25, 26]. Based on their inhibitory effects on cell proliferation, we analyzed the short-term effects of GADD45B and PMAIP1 overexpressions or knockdowns on cell growth of normal lung cells and the four human NSCLC cell lines using cell viability assays. It was observed that the number of viable cells was increased by TFAP2C-overexpressing WI-26 VA4 cells compared to that in control cells at day 5, and those of the viable cells in GADD45B or PMAIP1 siRNA-treated cells were considerably increased, nearly up to the level in the TFAP2C-overexpressing cells (Fig. 4a). In contrast, the number of survived cells in TFAP2C siRNA-treated NSCLC cells was lower, compared to control siRNA-treated cells, at day 5, and that of the survived cells was significantly rescued by further treatment of GADD45B or PMAIP1 siRNA (Fig. 4b–e). In addition, colony-forming assays were performed to determine whether the expressions of GADD45B and PMAIP1 regulated by TFAP2C were associated with long-term effects on cell proliferation. We found that the colony-forming capacity in WI-26 VA4 cells was enhanced by TFAP2C overexpression and GADD45B or PMAIP1 inhibition compared to that in control cells (Fig. 5a, b). In addition, we determined whether the expressions of TFAP2C, GADD45B, and PMAIP1 were associated with the apoptosis by examining the activation of caspase 3/7. The activities of caspase 3/7 in WI-26 VA4 cells were decreased by TFAP2C overexpression and GADD34B or PMAIP1 inhibition compared to control cells (Fig. 5c). Conversely, the colony formation ability in NCI-H292, NCI-H358, NCI-H460, and A549 cells was diminished, compared to that in control siRNA-treated cells, by treatment of TFAP2C siRNA, whereas colony formation was partially, but significantly, recovered by further GADD45B or PMAIP1 siRNA treatment (Fig. 5d, e). Moreover, the activities of caspase 3/7 in NSCLC cell lines were increased by TFAP2C siRNA treatment, compared to cells treated with control siRNA (Fig. 5f). The increased activities of caspase 3/7 by TFAP2C knockdown were reduced by further treatment of GADD45B or PMAIP1 siRNA. However, we observed that inhibition of GADD45B or PMAIP1 alone did not affect colony formation ability in four NSCLC cell lines compared to that in control cells (Additional file 1: Fig. S1). These results indicate that GADD45B and PMAIP1 expressions have important roles in NSCLC cell proliferation.
Short-term effects of TFAP2C, GADD45B, and PMAIP1 expression levels on NSCLC cell proliferation. a The effects of TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown on cell proliferation of WI-26 VA4 cells were measured by using cell viability assays. *p < 0.05 compared to cells treated with control vector or control siRNA. b–e The effects of TFAP2C, GADD45B, and/or PMAIP1 knockdown on cell proliferation of NCI-H292 (b), NCI-H358 (c), NCI-H460 (d), or A549 (e) cells were measured by using cell viability assays. *p < 0.05 compared to cells treated with control siRNA, **p < 0.05 compared to cells treated with TFAP2C siRNA
Long-term effects of TFAP2C, GADD45B, and PMAIP1 expression levels on NSCLC cell proliferation. a The effects of TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown on cell proliferation of WI-26 VA4 cells were measured using colony-forming assays. b Quantitative analysis of the number of WI-26 VA4 cell clones after TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown was performed. The colonies of TFAP2C-overexpressing cells set at 100%. *p < 0.05 compared to cells treated with control vector or control siRNA. c The effects of TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown on WI-26 VA4 cell apoptosis were determined using caspase 3/7 assays. *p < 0.05 compared to cells treated with control vector or control siRNA. d The effects of TFAP2C, GADD45B, and/or PMAIP1 knockdown on cell proliferation of NCI-H292, NCI-H358, NCI-H460, or A549 cells were measured by using colony-forming assays. Representative images of each group are presented. e Quantitative analysis of the number of four NSCLC cell clones after the treatments was performed. The colonies of non-treated control cells set at 100%. *p < 0.05 compared to cells treated with control siRNA, **p < 0.05 compared to cells treated with TFAP2C siRNA. f The effects of TFAP2C, GADD45B, and/or PMAIP1 knockdown on apoptosis of NCI-H292, NCI-H358, NCI-H460, or A549 cells were measured by using caspase 3/7 assays. *p < 0.05 compared to cells treated with control siRNA, **p < 0.05 compared to cells treated with TFAP2C siRNA
Increased cell motility in NSCLC cells by downregulation of GADD45B and PMAIP1 expression
Cell motility, which is highly correlated with epithelial-mesenchymal transition (EMT) invasion, and migration, is one of the common features related to cancer aggressiveness and malignant progression [27]. To determine the effects of GADD45B and PMAIP1 expression levels on cell motility, we conducted wound-healing assays and transwell cell migration assays in normal lung cells and NSCLC cells. We observed that WI-26 VA4 cells treated with TFAP2C overexpression, GADD45B siRNA, or PMAIP1 siRNA showed enhanced cell motility based on the time-dependent ratio of scratch coverage and migration capacity compared to those for control cells (Fig. 6a–c). In contrast, compared to those for control siRNA-treated cells, the covered areas and migrated cells for each of the four human NSCLC cell lines decreased with treatment of TFAP2C siRNA, and the uncovered areas were significantly filled and cell migration capacity was recovered after further treatment with GADD45B or PMAIP1 siRNA (Fig. 6d–f). The area-coverage tendencies, by cell line and based on repeated experiments, are depicted. However, we observed that inhibition of GADD45B or PMAIP1 alone did not affect wound-healing ability in four NSCLC cell lines compared to that in the control cells (Additional file 2: Fig. S2). Overall, these results indicate that the TFAP2C-suppressed expressions of GADD45B and PMAIP1 are positively involved in NSCLC malignancy including EMT, invasion, and migration.
The effects of TFAP2C, GADD45B, and PMAIP1 expression levels on NSCLC cell motility. a The effects of TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown on cell motility of WI-26 VA4 cells were measured by using wound-healing assays. Representative images of each group are presented. b The graphs show the percentage of the available area covered by WI-26 VA4 cells from five randomly selected images. Representative images of each group at different times are presented. *p < 0.05 compared to cells treated with control vector or control siRNA. c The effects of TFAP2C overexpression, GADD45B knockdown, or PMAIP1 knockdown on cell migration capacity of WI-26 VA4 cells were measured by using transwell cell migration assays. *p < 0.05 compared to cells treated with control vector or control siRNA. d The effects of TFAP2C, GADD45B, and/or PMAIP1 knockdown on cell motility of NCI-H292, NCI-H358, NCI-H460, or A549 cells were measured by using wound-healing assays. Representative images of each group are presented. e The graphs show the percentage of the available area covered by four NSCLC cell lines from five randomly selected images. *p < 0.05 compared to cells treated with control siRNA, **p < 0.05 compared to cells treated with TFAP2C siRNA. f The effects of TFAP2C, GADD45B, and/or PMAIP1 knockdown on cell migration capacity of NCI-H292, NCI-H358, NCI-H460, or A549 cells were measured by using transwell cell migration assays. *p < 0.05 compared to cells treated with control siRNA, **p < 0.05 compared to cells treated with TFAP2C siRNA
Discussion
In the current study, we investigated the involvements of TFAP2C and TFAP2C-regulated genes in lung tumorigenesis. Through an analysis of microarray data, we identified nine tumor suppressors, GADD45B, PMAIP1, XAF1, CYR61, IL24, ATF3, DLC1, RHOB and TNFAIP3, that were downregulated by TFAP2C overexpression in NSCLC cells. The low expression levels of the nine genes in lung cancer were confirmed by examination of the Oncomine data. Although all nine genes were involved in cancer cell death, tumor growth, angiogenesis, and EMT through various mechanisms [26, 28,29,30,31,32,33,34,35,36], we focused on GADD45B and PMAIP1 (also known as Noxa) as representative tumor suppressors in NSCLC tumorigenesis. Our results show that knockdowns of GADD45B and PMAIP1 conferred cell proliferation, colony-forming capacity, and cell motility on normal lung cells and NSCLC cells. Taken together, the results suggest that GADD45B and PMAIP1, when negatively regulated by TFAP2C, can act as tumor suppressive factors inhibiting NSCLC tumorigenesis.
GADD45B functions as a DNA damage sensor and has been studied as a positive regulator of apoptosis under various stimuli. Although GADD45B might be up-regulated and responsible for carcinogenesis under DNA damage response in a few cases [37, 38], many studies have presented that downregulation of GADD45B contributes to tumorigenesis and malignant development in human cancers. Another study reported that a loss of GADD45B was associated with upregulation of JNK and signal transducer and activator of transcription (STAT) 5, resulting in cell proliferation and inhibition of apoptosis in tumorigenesis of chronic myeloid leukemia [25]. In a study of functional analyses of chemotherapeutic agents, GADD45B was positively involved in G2/M-phase arrest and apoptosis through induction of cyclin-dependent kinase inhibitor 1A expression [39]. Overexpression of GADD45B has enhanced Fas-induced apoptosis through increased interaction of p38 with Rb protein [40]. In addition, the expression of GADD45B can be upregulated by Smad2, Smad3, and Smad4 signaling pathways stimulated by the transforming growth factor beta (TGF-β) signaling [41]. Another study demonstrated that STAT3 could bind to upstream regulatory elements of the GADD45B gene to function as a transcriptional repressor of GADD45B [42]. A recent study reported that the mRNA of STAT3 is targeted by miR-519a [43]. Interestingly, based our microarray dataset, we observed that miR-519a was slightly upregulated by TFAP2C knockdown. These clues suggest that low expression levels of GADD45B might be indirectly due to TFAP2C-mediated downregulation of miR-519a, although we could not exclude the possibility that TFAP2C can act as a direct repressor of GADD45B.
PMAIP1 belongs to the BH3-only pro-apoptotic protein of the Bcl-2 family and upregulation of PMAIP1 leads to apoptosis in a variety of cancers. Downregulation of PMAIP1 has been shown to promote the expression of ubiquitin-specific peptidase 9, X-linked, and myeloid cell leukemia 1, resulting in apoptosis in NSCLC cells [26]. In addition, it has been reported that PMAIP1 has an important role in the induction of apoptosis and autophagy, and was shown to be downregulated in adenoid cystic carcinoma [44]. Another study demonstrated that PMAIP1 mRNA can be targeted by miR-21 in gastric carcinoma, resulting in the inhibition of proliferation, invasion, and migration [45]. In particular, a study using chromatin immunoprecipitation showed that activating transcription factor (ATF) 3 and ATF4 could be putative transcription factors responsible for the expression of PMAIP1 [46]. That suggestion was supported by another study in which cisplatin-induced ATF3 in head and neck squamous cell carcinoma cells resulted in induction of apoptosis accompanied by an increased level of PMAIP1 through cooperative binding of ATF3 and ATF4 to the PMAIP1 promoter [47]. As shown in Figs. 1 and 2, we also found ATF3 to be upregulated by TFAP2C knockdown, based on the analysis of the TFAP2C-related microarray dataset. Although further molecular studies into the interplay between TFAP2C, ATF3, and PMAIP1 would be required, we suggest that oncogenic transcription factor TFAP2C might contribute to NSCLC tumorigenesis by downregulation of PMAIP1 in an ATF3-dependent manner.
Conclusion
There have been many studies seeking to identify molecular targets useful in elucidating the molecular mechanisms underlying tumorigenesis and malignant development of NSCLC. The results of the present study have shown that TFAP2C can contribute to NSCLC tumorigenesis via downregulation of many tumor suppressors including GADD45B, PMAIP1, XAF1, CYR61, IL24, ATF3, DLC1, RHOB, and TNFAIP3. For the first time, we report that GADD45B and PMAIP1, which are suppressed by TFAP2C, could be highly responsible for cell proliferation and cell motility in NSCLC. We propose that GADD45B and PMAIP1 be considered putative tumor suppressive factors in NSCLC that might be useful as prognostic markers during NSCLC therapy.
Availability of data and materials
All data or data analyzed during this study are included in this published article (and its additional information files).
Abbreviations
- NSCLC:
-
non-small cell lung cancer
- GADD45B:
-
growth arrest and DNA-damage-inducible beta
- PMAIP1:
-
phorbol-12-myristate-13-acetate-induced protein 1
- TFAP2C:
-
transcription factor activating enhancer-binding protein 2C
- RT:
-
reverse transcription
- SD:
-
standard deviation
- STAT:
-
signal transducer and activator of transcription
- ATF:
-
activating transcription factor
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
O’Shea K, Cameron SJ, Lewis KE, et al. Metabolomic-based biomarker discovery for non-invasive lung cancer screening: a case study. Biochim Biophys Acta. 2016;1860:2682–7.
Eckert D, Buhl S, Weber S, et al. The AP-2 family of transcription factors. Genome Biol. 2005;6:246.
Lal G, Contreras PG, Kulak M, et al. Human melanoma cells over-express extracellular matrix 1 (ECM1) which is regulated by TFAP2C. PLoS ONE. 2013;8:e73953.
De Andrade JP, Park JM, Gu VW, et al. EGFR is regulated by TFAP2C in luminal breast cancer and is a target for vandetanib. Mol Cancer Ther. 2016;15:503–11.
Kang J, Kim W, Lee S, et al. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183- and miR-33a-mediated cell cycle regulation. Oncogene. 2017;36:1585–96.
Kim W, Kim E, Lee S, et al. TFAP2C-mediated upregulation of TGFBR1 promotes lung tumorigenesis and epithelial–mesenchymal transition. Exp Mol Med. 2016;48:e273.
Wang X, Sun D, Tai J, et al. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J Exp Clin Cancer Res. 2018;37:27.
Kang J, Kim W, Seo H, et al. Radiation-induced overexpression of transthyretin inhibits retinol-mediated hippocampal neurogenesis. Sci Rep. 2018;8:8394.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8.
Kim W, Youn H, Lee S, et al. RNF138-mediated ubiquitination of rpS3 is required for resistance of glioblastoma cells to radiation-induced apoptosis. Exp Mol Med. 2018;50:e434.
Kim E, Kim W, Lee S, et al. TRAF4 promotes lung cancer aggressiveness by modulating tumor microenvironment in normal fibroblasts. Sci Rep. 2017;7:8923.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Kim W, Youn H, Kang C, et al. Inflammation-induced radioresistance is mediated by ROS-dependent inactivation of protein phosphatase 1 in non-small cell lung cancer cells. Apoptosis. 2015;20:1242–52.
Son B, Kwon T, Lee S, et al. CYP2E1 regulates the development of radiation-induced pulmonary fibrosis via ER stress- and ROS-dependent mechanisms. Am J Physiol Lung Cell Mol Physiol. 2017;313:L916–29.
Li J, Tan Q, Yan M, et al. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166.
Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6.
Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–5.
Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93.
Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5:e10312.
Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–211.
Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3:e1651.
Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–11.
Sha X, Hoffman B, Liebermann DA. Loss of Gadd45b accelerates BCR-ABL-driven CML. Oncotarget. 2018;9:33360–7.
Yan J, Zhong N, Liu G, et al. Usp9x- and Noxa-mediated Mcl-1 downregulation contributes to pemetrexed-induced apoptosis in human non-small-cell lung cancer cells. Cell Death Dis. 2014;5:e1316.
Lu W, Kang Y. Epithelial–mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49:361–74.
Jiang X, Yin L, Zhang N, et al. Bisphenol A induced male germ cell apoptosis via IFNbeta-XAF1-XIAP pathway in adult mice. Toxicol Appl Pharmacol. 2018;355:247–56.
Feng P, Wang B, Ren EC. Cyr61/CCN1 is a tumor suppressor in human hepatocellular carcinoma and involved in DNA damage response. Int J Biochem Cell Biol. 2008;40:98–109.
Chen QN, Chen X, Chen ZY, et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer. 2017;16:17.
Tian Z, An N, Zhou B, et al. Cytotoxic diarylheptanoid induces cell cycle arrest and apoptosis via increasing ATF3 and stabilizing p53 in SH-SY5Y cells. Cancer Chemother Pharmacol. 2009;63:1131–9.
Yang X, Zhou X, Tone P, et al. Cooperative antiproliferative effect of coordinated ectopic expression of DLC1 tumor suppressor protein and silencing of MYC oncogene expression in liver cancer cells: therapeutic implications. Oncol Lett. 2016;12:1591–6.
Du X, Qian X, Papageorge A, et al. Functional interaction of tumor suppressor DLC1 and caveolin-1 in cancer cells. Cancer Res. 2012;72:4405–16.
Bousquet E, Calvayrac O, Mazieres J, et al. RhoB loss induces Rac1-dependent mesenchymal cell invasion in lung cells through PP2A inhibition. Oncogene. 2016;35:1760–9.
Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–41.
Michaelis KA, Knox AJ, Xu M, et al. Identification of growth arrest and DNA-damage-inducible gene beta (GADD45beta) as a novel tumor suppressor in pituitary gonadotrope tumors. Endocrinology. 2011;152:3603–13.
Jin X, Liu X, Zhang Z, et al. Identification of key pathways and genes in lung carcinogenesis. Oncol Lett. 2018;16:4185–92.
Wang L, Xiao X, Li D, et al. Abnormal expression of GADD45B in human colorectal carcinoma. J Transl Med. 2012;10:215.
Nettersheim D, Jostes S, Fabry M, et al. A signaling cascade including ARID1A, GADD45B and DUSP1 induces apoptosis and affects the cell cycle of germ cell cancers after romidepsin treatment. Oncotarget. 2016;7:74931–46.
Cho HJ, Park SM, Hwang EM, et al. Gadd45b mediates Fas-induced apoptosis by enhancing the interaction between p38 and retinoblastoma tumor suppressor. J Biol Chem. 2010;285:25500–5.
Yoo J, Ghiassi M, Jirmanova L, et al. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278:43001–7.
Kim JH, Qu A, Reddy JK, et al. Hepatic oxidative stress activates the Gadd45b gene by way of degradation of the transcriptional repressor STAT3. Hepatology. 2014;59:695–704.
Tian F, Jia L, Chu Z, et al. MicroRNA-519a inhibits the proliferation and promotes the apoptosis of ovarian cancer cells through targeting signal transducer and activator of transcription 3. Exp Ther Med. 2018;15:1819–24.
Liang L, Weng J, You Y, et al. Role of Noxa in proliferation, apoptosis, and autophagy in human adenoid cystic carcinoma. J Oral Pathol Med. 2019;48:52–9.
Sun H, Wang P, Zhang Q, et al. MicroRNA21 expression is associated with the clinical features of patients with gastric carcinoma and affects the proliferation, invasion and migration of gastric cancer cells by regulating Noxa. Mol Med Rep. 2016;13:2701–7.
Narita T, Ri M, Masaki A, et al. Lower expression of activating transcription factors 3 and 4 correlates with shorter progression-free survival in multiple myeloma patients receiving bortezomib plus dexamethasone therapy. Blood Cancer J. 2015;5:e373.
Sharma K, Vu TT, Cook W, et al. p53-independent Noxa induction by cisplatin is regulated by ATF3/ATF4 in head and neck squamous cell carcinoma cells. Mol Oncol. 2018;12:788–98.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03932822) and the Ministry of Science and ICT (2019R1C1C1009423).
Author information
Authors and Affiliations
Contributions
HD, BY and WK designed study strategy; HD, DK, DS, and WK performed experiments; HD, DK, JK, BS, DS, HY, BY, and WK analyzed the data; HD, DK, BY and WK wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Fig. S1.
The effects of TFAP2C, GADD45B and PMAIP1 expression levels on NSCLC cell proliferation.
Additional file 2: Fig. S2.
The effects of TFAP2C, GADD45B and PMAIP1 expression levels on NSCLC cell motility.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Do, H., Kim, D., Kang, J. et al. TFAP2C increases cell proliferation by downregulating GADD45B and PMAIP1 in non-small cell lung cancer cells. Biol Res 52, 35 (2019). https://doi.org/10.1186/s40659-019-0244-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40659-019-0244-5