Abstract
Background
Acute myeloid leukemia (AML) with the t(8;21)(q22;q22) chromosomal translocation is among the most common subtypes of AML and produces the AML1-ETO (RUNX1-ETO, RUNX1-RUNX1T1) oncogenic fusion gene. AML1-ETO functions as an aberrant transcription factor which plays a key role in blocking normal hematopoiesis. Thus, the expression of AML1-ETO is critical to t(8;21) AML leukemogenesis and maintenance. Post-transcriptional regulation of gene expression is often mediated through interactions between trans-factors and cis-elements within transcript 3′-untranslated regions (UTR). AML1-ETO uses the 3′UTR of the ETO gene, which is not normally expressed in hematopoietic cells. Therefore, the mechanisms regulating AML1-ETO expression via the 3’UTR are attractive therapeutic targets.
Methods
We used RNA-sequencing of t(8;21) patients and cell lines to examine the 3′UTR isoforms used by AML1-ETO transcripts. Using luciferase assay approaches, we test the relative contribution of 3′UTR cis elements to AML1-ETO expression. We further use let-7b microRNA mimics and anti-let-7b sponges for functional studies of t(8;21) AML cell lines.
Results
In this study, we examine the regulation of AML1-ETO via the 3’UTR. We demonstrate that AML1-ETO transcripts primarily use a 3.7 kb isoform of the ETO 3′UTR in both t(8;21) patients and cell lines. We identify a negative regulatory element within the AML1-ETO 3′UTR. We further demonstrate that the let-7b microRNA directly represses AML1-ETO through this site. Finally, we find that let-7b inhibits the proliferation of t(8;21) AML cell lines, rescues expression of AML1-ETO target genes, and promotes differentiation.
Conclusions
AML1-ETO is post-transcriptionally regulated by let-7b, which contributes to the leukemic phenotype of t(8;21) AML and may be important for t(8;21) leukemogenesis and maintenance.
Similar content being viewed by others
Introduction
The t(8;21)(q22;q22) translocation is among the most common chromosomal abnormalities in AML [1]. Although t(8;21) AML is categorized as a favorable-risk AML, there remains a high incidence of relapse (up to 56 % in some studies) and the median overall survival is only 5 years, highlighting the need for further studies and additional therapies [1,2,3,4,5]. The t(8;21) translocation results in a fusion of the AML1 (RUNX1) locus on chromosome 21 with the ETO (MTG8, RUNX1T1) locus on chromosome 8, creating the AML1-ETO (RUNX1-ETO, RUNX1-RUNX1T1) fusion gene [6]. AML1 is an essential transcription factor for healthy hematopoiesis [7]. The N-terminal runt homology domain (RHD) of AML1 mediates sequence-specific DNA-binding whereas the C-terminal transcriptional regulatory domain controls target gene transcription through the recruitment of essential co-factors [7]. ETO (RUNX1T1) is not normally expressed in hematopoietic progenitor cells [8,9,10], does not have DNA binding activity [11], and interacts with transcriptional co-repressors and co-activators [6, 11,12,13,14,15,16]. The AML1-ETO chimeric transcript contains the N-terminal RHD of AML1 and nearly the entire ETO gene, including its 3′UTR [6]. Thus, the AML1-ETO fusion transcription factor impairs normal AML1-mediated myeloid differentiation through the transcriptional dysregulation of critical hematopoietic genes [6, 17,18,19,20,21,22,23,24,25], although additional mutations are also required for leukemogenesis [26, 27].
Stable expression of the AML1-ETO fusion gene is critical for both the initiation and maintenance of t(8;21) AML [28,29,30,31,32,33,34]. Transient knockdown of AML1-ETO induces differentiation [28, 30], inhibits proliferation [29, 30], and reduces leukemic burden [31]. AML1 expression is required for healthy hematopoiesis [7]. In contrast, ETO is not normally expressed in healthy hematopoietic progenitor cells [8,9,10]. Therefore, the factors which regulate AML1-ETO expression through the ETO portion of the chimeric transcript, including the 3’UTR, may be a unique sensitivity of t(8;21) leukemia cells compared to healthy hematopoietic cells.
The 3’UTR plays a crucial role in post-transcriptional gene expression by influencing the localization, stability, export, and translation efficiency of mRNA transcripts [35]. Sequence specific interactions between cis-elements within the 3′UTR and trans-factors, such as RNA binding proteins or microRNAs (miRNA), are major mediators of post-transcriptional regulation [35, 36]. miRNAs have also been shown to promote or suppress a variety of leukemic processes, including proliferation, apoptosis, differentiation, self-renewal, epigenetic regulation, and chemotherapy resistance [37,38,39]. The let-7 family of miRNAs are well described tumor suppressors in a variety of cancers, including leukemias [40,41,42]. Members of the let-7 miRNA family have demonstrated roles in sensitizing leukemic cells to chemotherapy [43,44,45,46,47]. Furthermore, the overexpression of let-7 miRNAs have also been shown to inhibit proliferation and promote differentiation in certain leukemic contexts [40, 48].
Since AML1-ETO expression is critical for t(8;21) AML leukemia, identifying factors that post-transcriptionally regulate AML1-ETO via the ETO 3′UTR may improve our understanding of how AML1-ETO expression is maintained and uncover suitable targets for t(8;21) AML therapy. Here, we demonstrate that AML1-ETO transcripts predominantly use the first 3.7 kb of the ETO 3′UTR, in both t(8;21) AML patients and cell lines. Using a luciferase assay approach to identify regulatory elements within the 3.7 kb AML1-ETO 3′UTR, we found a fragment of the AML1-ETO 3′UTR between 2.8 and 3.4 kb which was negatively regulated, increased expression upon inhibition of miRNA biogenesis, and contained a putative miRNA let-7 target site. We further showed that let-7b targets and represses AML1-ETO expression through this site. Finally, exogenous let-7b miRNA transfection inhibited the proliferation of t(8;21) AML cell lines, rescued expression of AML1-ETO downregulated target genes, and promoted leukemia cell differentiation. Our findings establish that the let-7b miRNA is a post-transcriptional regulator of AML1-ETO and affects the leukemic phenotype of t(8;21) AML.
Methods
Patients samples
t(8;21) AML samples were obtained from patients at UC San Diego Health. Following collection of peripheral blood or bone marrow from AML patients, cells were separated using Ficoll-Paque (VWR, Radnor, PA) and frozen until further use. Thawed products were diluted in 1x PBS supplemented with 1 mg/mL DNAse I (Sigma-Aldrich, St. Louis, MO) and washed with 1x PBS supplemented with 2 % FBS. Live, mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque (VWR). Magnetic bead CD34-enrichment was performed using a human CD34 MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany), following the manufacturer’s instructions. An aliquot of CD34-enriched leukemic blasts was analyzed by flow cytometry to confirm that cells were > 95 % CD34+. RNA was extracted from patient blasts using Trizol reagent (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions.
CD34+ HSPCs from healthy donors were obtained from Fred Hutchinson Cooperative Center for Excellence in Hematology (Seattle, Washington). Cells were thawed quickly and serially diluted with 1x PBS supplemented with 2 % FBS. Cells were resuspended in Trizol for RNA extraction.
RNA-sequencing
Total RNA from Kasumi-1 and SKNO-1 cells was isolated using Trizol reagent (ThermoFisher). Library preparation of total RNA from Kasumi-1, SKNO-1, healthy HSPCs and patient blasts was performed using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA) followed by sequencing on an Illumina HiSeq4000. Available public data for normal brain samples (SRR5938419 and SRR5938420) was downloaded from the NCBI Gene Expression Omnibus database [49]. Raw RNA-seq reads were aligned and mapped using HISAT2 using the usegalaxy public supercomputing platform [50].
Transfection
One to two million cells of THP-1, HL-60, Kasumi-1, SKNO-1, or associated cell lines were resuspended with plasmid DNA or miRNA mimics in 100 µL of transfection buffer [140 mM Na2HPO4/NaH2PO4 (pH 7.2), 5 mM KCl, 15 mM MgCl2]. Cells were then transfected using programs P-19 (SKNO-1 and Kasumi-1), U-01 (THP-1), or Y-01 (HL-60) of the AMAXA II Nucleofector (Lonza; Basel, Switzerland) and were cultured in RPMI media plus 20 % FBS in a 37°C incubator with 5 % CO2. After 24 h the media was changed to RPMI plus 10 % FBS and 100 U/mL penicillin-streptomycin for the indicated times. For the HEK293T reporter assay, HEK293T cells were plated in 96-well plates and transfected with 5 ng of the psiCHECK2 3.7 kb AML1-ETO 3’UTR reporter plasmid DNA and 10 pmol of miRIDIAN™ microRNA Mimic negative control #2 or indicated miRIDIAN™ microRNA mimics (Dharmacon, Lafayette, CO), using DharmaFECT™ Duo transfection reagent (Dharmacon) according to manufacturer’s instructions.
Proliferation assay
miRIDIAN™ microRNA Mimic negative control #2 or hsa-let-7b-5p (Dharmacon) were transfected into the SKNO-1 (100 pmol), THP-1 (100 pmol), HL-60 (200 pmol), or Kasumi-1 (200 pmol) cell lines. Viable cells were counted via trypan blue exclusion using a TC20 Automated Cell Counter (Bio-Rad Laboratories, Hercules, CA) on indicated days and were initially cultured at a density of 2 × 105 cells per mL.
Western blot
Primary antibodies included a mouse anti-α-tubulin (1:10,000) antibody (12G10, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and a previously described rabbit anti-AML1 (1:500) antibody generated by Covance[23]. Licor (Lincoln, NE) IRDye 680RD goat anti-mouse IgG and IRDye 800CW goat anti-rabbit IgG secondary antibodies (1:10,000) were used for visualization on a LI-COR Odyssey Classic imager. Image analysis and densitometry were performed using the LI-COR Application Software Version 3.0.
Statistical analyses
All statistical analyses were performed using GraphPad Prism Software (Version 8.4.2). The specific tests used are documented in the corresponding figure legend. All t-tests were two-tailed. P values are denoted as follows: ns p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001.
Results
AML1-ETO transcripts primarily use the first 3.7 kb of the ETO 3′UTR in t(8;21) AML patients and cell lines
To uncover post-transcriptional regulators of the AML1-ETO oncogene that target the ETO 3’UTR, we first sought to identify which regions of the 3’UTR are present in AML1-ETO transcripts. Alternative polyadenylation regulates 3’UTR length and consequently the availability of cis-acting elements that bind trans-acting post-transcriptional regulators [35]. Therefore, to identify t(8;21) AML relevant interactions, we analyzed the endogenous AML1-ETO 3’UTR usage in t(8;21) AML patients and cell lines.
We performed RNA-seq on peripheral blood derived CD34+ HSPCs from healthy donors and CD34+ enriched t(8;21) AML primary patients’ blasts. The longest predicted AML1-ETO 3′UTR sequence is 5.2 kb in length. However, analysis of the sequencing reads in the t(8;21) patients showed that the abundance of reads aligned within the first 3.7 kb of the ETO 3′UTR and only minor signal is detected up to the full 5.2 kb (Fig. 1a). Importantly, a validated poly(A) site is located at 3.7 kb, suggesting that AML1-ETO transcripts terminate at this proximal poly(A) site resulting in the observed shorter 3′UTR in patients. Consistent with previous reports [8,9,10], wild type ETO expression was undetectable in the healthy HSPC samples and the ETO RNA-seq reads in t(8;21) patient samples only map to the exons present in the AML1-ETO fusion (Fig. 1a, Additional file 1: S1a, b) [6]. To compare AML1-ETO 3′UTR usage to wild type ETO in a normal physiological context, we analyzed publicly available RNA-seq data of healthy brain cortex [49], which is known to express ETO [10]. The normal brain cortex RNA-seq reads included peaks in exons not involved in the AML1-ETO fusion and had a very similar pattern within the ETO 3′UTR, with the abundance of reads mapping within the first 3.7 kb of the ETO 3′UTR (Additional file 1: Fig. S1a, b). We further performed RNA-seq using the t(8;21) AML cell lines Kasumi-1 and SKNO-1. Interestingly, nearly all the AML1-ETO 3′UTR reads aligned within the first 3.7 kb in these cell lines (Fig. 1b). Taken together, our data suggests that AML1-ETO fusion transcripts primarily use the first 3.7 kb of the ETO 3′UTR in t(8;21) patients and cell lines.
AML1-ETO transcripts primarily use a 3.7 kb isoform of the ETO 3′UTR in t(8;21) AML patients and cell lines. a Alignment of RNA-seq reads from healthy HSPCs (n = 2) and t(8;21) AML patient blasts (n = 4) to the final exon of ETO. Putative 3.7 kb and full-length 5.2 kb ETO 3′UTR isoforms are depicted below. b Alignment of RNA-seq reads from the t(8;21)+ Kasumi-1 and SKNO-1 cell lines to the final exon of ETO. Putative 3.7 kb and full-length 5.2 kb ETO 3′UTR isoforms are depicted below
AML1-ETO 3′UTR cis-elements affect expression and are targeted by miRNAs
We next sought to identify the relevant regulatory sequences within the AML1-ETO 3′UTR. Because AML1-ETO patient transcripts predominantly used the first 3.7 kb of the ETO 3′UTR, we focused on sequences within this region. To identify regions within the 3’UTR that contain cis-elements that regulate AML1-ETO expression, we created a set of dual luciferase reporters that express Renilla luciferase with 3′UTRs containing progressive 600 bp fragments spanning the length of the first 3.7 kb of the ETO 3′UTR (with 200 bp overlaps between fragments) and firefly luciferase as an internal control (Fig. 2a). We mapped the effects of the AML1-ETO 3′UTR fragments on luciferase reporter expression compared to a control reporter through luciferase assays after transient transfection into the t(8;21)+ Kasumi-1 and SKNO-1 cell lines (Fig. 2b). The reporters containing AML1-ETO 3′UTR fragments had equal or lower expression compared to the control reporter, with some having up to 9-fold lower expression. We also observed up to 7-fold differences in luciferase activity between the reporters containing AML1-ETO 3′UTR fragments. Interestingly, reporters containing fragments near the 3’ end of the AML1-ETO 3′UTR (#7 2400–3000 bp, #8 2800–3400 bp, and #9 3100–3741 bp) had the lowest expression levels in both Kasumi-1 and SKNO-1 cells, suggesting that there are negatively regulated cis-elements in this region.
AML1-ETO 3′UTR cis-elements affect expression and are targeted by miRNAs. a Schematic of the dual luciferase reporters containing 600 bp fragments of the AML1-ETO 3′UTR. b Individual AML1-ETO 3′UTR dual luciferase reporters were transfected into Kasumi-1 and SKNO-1 cells and luciferase activity was measured. Assay results were normalized to an empty vector control. Values and error bars represent the average and SD of relative luciferase from three independent experiments. Significance is shown compared to empty vector control using one-way ANOVA with a post-hoc Tukey test. c Representative western blot and quantification of AML1-ETO protein in SKNO-1 DICER knockdown or control shRNA cell lines. Values and error bars represent the mean and SD of three independent experiments. d SKNO-1 cells expressing shRNAs targeting DICER or control shRNA were transfected with dual luciferase reporters expressing the indicated AML1-ETO 3′UTR fragments. Values and error bars represent the mean and SD of relative luciferase activity of shDICER compared to shCTRL from indicated number of independent experiments. Significance is shown compared to shCTRL control using unpaired t-tests corrected for multiple comparisons using the Holm-Sidak method
miRNAs are major mediators of negative post-transcriptional regulation. Normal miRNA biogenesis requires a multi-step process involving the RNAse DICER [51]. To assess the contribution of miRNAs in the regulation of AML1-ETO, we made stable shRNA-mediated DICER knockdown or control shRNA transduced SKNO-1 cell lines. We observed significantly increased levels of AML1-ETO protein upon DICER knockdown, compared with controls (Fig. 2c, Additional file 1: S2a). This result suggests that miRNAs regulate AML1-ETO protein levels and supports previous work that identified miR-193a, miR-9, and miR-29b as AML1-ETO regulators [52,53,54]. However, none of these miRNAs had predicted target sites within the 2600-3741 bp negatively regulated fragments near the end of the AML1-ETO 3’UTR [52,53,54]. To determine if miRNAs were involved specifically in the negative regulation of our AML1-ETO 3′UTR reporters, we transfected the lowest expressing 600 bp AML1-ETO 3′UTR reporters into the SKNO-1 shRNA control and shRNA DICER cell lines and performed luciferase assays (Fig. 2d). We observed significantly increased luciferase activity in both DICER knockdown cell lines transfected with the reporter containing the AML1-ETO 3’UTR fragment #8 (2800–3400 bp). Together, these results suggest that miRNAs are involved in the regulation of AML1-ETO and that at least one such miRNA specifically targets the AML1-ETO 3′UTR between 2800 and 3400 bp.
let-7b targets and down regulates AML1-ETO via the 3’UTR
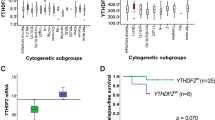
To find miRNAs which putatively target the AML1-ETO 3’UTR between 2800 and 3400 bp, we used a miRNA targeting prediction model, TargetScan 7.2 [55]. We identified 6 families of miRNAs that were predicted to target this region: let-7, miR-33, miR-129, miR-153, miR-190, and miR-202 (Fig. 3a). We next tested the ability of all 6 candidate miRNA families to downregulate the AML1-ETO 3’UTR. We co-transfected miRNA “mimics” or non-targeting control mimics with a luciferase reporter containing the full 3.7 kb AML1-ETO 3’UTR in HEK293T cells (Fig. 3b). The let-7 family member mimic, let-7b, significantly decreased luciferase activity of the AML1-ETO 3′UTR reporter, suggesting that let-7b targets the AML1-ETO 3’UTR. Interestingly, among favorable-risk AML patients in the TCGA adult AML dataset [56], those with high expression of let-7b had significantly higher overall and disease-free survival (Additional file 1: Fig. S3a); though direct correlation between let-7b expression and overall and disease-free survival was only weakly positive (Additional file 1: Fig. S3b). Based on the potential survival benefit and the AML1-ETO 3’UTR reporter results, we focused on let-7b for our further studies.
let-7b targets and down regulates an AML1-ETO 3’UTR reporter in HEK293T cells. a Schematic representation of miRNAs with predicted target sites within the 2800–3400 bp AML1-ETO 3′UTR fragment. Predictions were made using TargetScan 7.2. b miRNA mimics or non-targeting control mimic were co-transfected into HEK293T cells with the AML1-ETO 3.7 kb 3′UTR luciferase reporter and luciferase assays were performed 24 h post-transfection. Data is shown as relative luciferase activity normalized to non-targeting control mimic. Significance was determined using one-way ANOVA with a post-hoc Tukey test
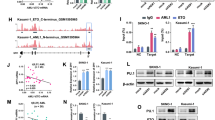
We investigated the effect of let-7b on regulating endogenous AML1-ETO by transfecting let-7b-5p or non-targeting control miRNA mimics into the Kasumi-1 and SKNO-1 cell lines (Fig. 4a, Additional file 1: S4a). We observed significantly decreased AML1-ETO protein expression in both Kasumi-1 and SKNO-1 cells that were transfected with the let-7b-5p mimics compared to controls (Fig. 4a). To further test the role of let-7 miRNAs in regulating endogenous AML1-ETO expression, we made stable Kasumi-1 and SKNO-1 cell lines which expressed an anti-let-7 family member miRNA sponge [57]. Consistent with previous reports [58], we observed a modest decrease in mature let-7b-5p levels in the anti-let-7 miRNA sponge expressing cell lines because the major activity of bulged miRNA sponges is through miRNA decoy rather than miRNA decay (Additional file 1: Fig. S4b). Both the Kasumi-1 and SKNO-1 cell lines which stably expressed let-7 miRNA sponges showed significantly increased AML1-ETO protein levels compared to control cell lines (Fig. 4b). Together, these results demonstrate that AML1-ETO protein is down-regulated upon the addition of exogenous let-7b miRNA and is up-regulated when endogenous let-7 family members are inhibited.
let-7b targets the 3’UTR and downregulates AML1-ETO in t(8;21) AML cells. a, b Representative western blots and protein quantification of Kasumi-1 and SKNO-1 whole cell lysates probed for endogenous AML1-ETO or α-Tubulin 48 h post-transfection with let-7b-5p or control miRNA mimics (a), or in cells stably expressing an anti-let-7 miRNA family sponge or control cells (b). c let-7b-5p or control miRNA mimics were co-transfected into Kasumi-1 cells with the AML1-ETO 3′UTR fragment #8 luciferase reporter and luciferase assays were performed 72 h post-transfection. Data is shown as relative luciferase activity normalized to control mimics. d Schematic of WT (AE-WT) and let-7 targeting mutant (AE-MUT) AML1-ETO 3′UTR fragment #8 luciferase reporters. e Luciferase assay results of AE-WT and AE-MUT reporters in Kasumi-1 or SKNO-1 cells 72 h post-transfection. f Kasumi-1 and SKNO-1 cell lines stably expressing either an anti-let-7 miRNA family sponge or control cells were transfected with the AE-WT or AE-MUT 3′UTR reporters and luciferase assays were performed 72 h later. Data is shown as relative luciferase activity normalized to control cell lines. All data in this figure is presented as the average and SD of the indicated number of individual experiments, unpaired t-tests with corrections for multiple tests using the Holm-Sidak method
The AML1-ETO 3’UTR contains a seed sequence for let-7 miRNA family members at 2.85 kb. Therefore, we next examined whether the regulation of AML1-ETO expression by let-7b was due to direct targeting of the AML1-ETO 3′UTR at this site. Indeed, luciferase activity of the AML1-ETO 3′UTR fragment #8 (2,800-3,400 bp) luciferase reporter was significantly decreased in Kasumi-1 cells co-transfected with let-7b-5p miRNA mimics compared to non-targeting control (Fig. 4c). Next, we introduced mutations of the putative let-7 target site (AE-MUT 3‘UTR) within the wild-type AML1-ETO 3’UTR #8 fragment luciferase reporter (AE-WT 3′UTR) (Fig. 4d). Luciferase assays in Kasumi-1 and SKNO-1 cells showed significantly increased activity in the reporters with the mutated let-7 target site compared to wild-type (Fig. 4e). Luciferase activity of the AE-WT 3′UTR reporter was also significantly greater in let-7 sponge expressing SKNO-1 and Kasumi-1 lines versus controls, whereas there was no significant difference in luciferase production by the AE-MUT 3′UTR reporter in these cellular contexts (Fig. 4f). Together, these results suggest that let-7b directly targets and down-regulates AML1-ETO via the 3′UTR.
Expression of let-7b inhibits cell growth in t(8;21) AML cell lines
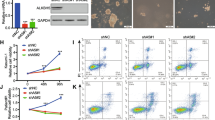
Having observed that let-7b-5p overexpression can reduce AML1-ETO protein expression, we next wondered whether the degree of AML1-ETO downregulation has potential therapeutic relevance. Therefore, we first measured the expression of known transcriptionally repressed AML1-ETO target genes, CEBPA [17] and RASSF2 [59], upon transfection of Kasumi-1 and SKNO-1 cells with the let-7b-5p miRNA mimic. In both cell lines, treatment with the miRNA mimic resulted in a significant increase of both transcripts in comparison to cells treated with a non-targeting control mimic (Fig. 5a). These results suggest that down regulation of AML1-ETO by let-7b-5p can partially rescue expression of genes that are transcriptionally repressed directly by AML1-ETO. We next measured the phenotypic impact of let-7 restoration on t(8;21)+ AML cell lines. The AML1-ETO protein has been shown to impair myeloid differentiation and transiently silencing its expression reduces cell proliferation and induces differentiation [24, 25, 28,29,30]. Likewise, the restoration of let-7 expression has been shown to inhibit cell proliferation and induce differentiation in cancers with low let-7 expression [60, 61]. Indeed, we observed significant inhibition of cell proliferation in the Kasumi-1 and SKNO-1 t(8;21) AML cell lines, but not in the HL-60 or THP-1 non-t(8;21) AML cell lines, after transfection with the let-7b-5p miRNA mimic versus the non-targeting control mimic (Fig. 5b). Additionally, both Kasumi-1 and SKNO-1 cells treated with the let-7b-5p mimic had significantly reduced cell surface expression of the hematopoietic stem cell marker, CD34 (Fig. 5 c, d). Furthermore, let-7b-5p mimic treated cells had significantly increased cell surface expression of the CD38 progenitor marker, as well as the CD33 and CD13 myeloid markers (Fig. 5 c, d). These results are indicative of a more differentiated cellular state, consistent with release of the AML1-ETO mediated myeloid differentiation block. Collectively, these results demonstrate that the restoration of let-7b levels rescues expression of repressed AML1-ETO target genes, impairs t(8;21) AML cell proliferation, and induces differentiation.
Expression of let-7b inhibits cell growth in t(8;21) AML cell lines. a Either let-7b-5p or control miRNA mimics were transfected into Kasumi-1 or SKNO-1 cells at an amount of 200 or 100 pmol for each respective cell line. Relative expression of CEBPA and RASSF2 were determined by qPCR 96 h post-transfection. Data is presented as let-7b-5p mimic fold change relative to control. Values represent mean and SD of three individual experiments. Significance was determined by unpaired t-tests. b Cell growth of the t(8;21) AML cell lines Kasumi-1 and SKNO-1 or the non-t(8;21) AML cell lines HL-60 and THP-1, after transfection with let-7b-5p or control miRNA mimics, was determined by cell counting on the indicated days. Values represent mean and SD of four individual experiments. Significance was determined by unpaired t-tests of individual timepoints assuming homoscedasticity and with a correction for multiple tests using the Holm-Sidak method. c, d Relative mean fluorescent intensity (MFI) of CD34, CD33, CD38, and CD13 in Kasumi-1 or SKNO-1 96 h post-transfection with let-7b-5p or control miRNA mimics was determined using flow-cytometry. Values represent MFI relative to control mimic from indicated number of individual experiments. Significance determined by unpaired t-tests corrected for multiple comparisons using the Holm-Sidak method. d Representative histograms depict the fluorescence distribution of indicated markers in the indicated cell populations
Discussion
The AML1-ETO oncofusion protein impairs myeloid differentiation and is important for the initiation and maintenance of t(8;21) AML [24, 25, 28,29,30,31,32,33,34]. Thus, AML1-ETO 3′UTR is an appropriate target for t(8;21) AML therapies due to importance of AML1-ETO expression to t(8;21) AML and the lack of wild type ETO expression in healthy hematopoietic cells. However, the post-transcriptional regulation of AML1-ETO via its 3′UTR has not been well studied. We demonstrate that AML1-ETO transcripts primarily use a 3.7 kb isoform which is targeted and repressed by the let-7b miRNA. Furthermore, we show that the transient expression of let-7b in t(8;21) AML cell lines confers a tumor-suppressive phenotype, partially rescues AML1-ETO target gene expression, and induces differentiation.
Our study is the first examination of AML1-ETO 3′UTR usage in t(8;21) AML patients and cell lines. Interestingly, the 3′UTR usage within our set of four t(8;21) AML patient samples was quite similar, with the majority of RNA-seq reads mapping within the first 3.7 kb of the 3′UTR. Based on these results, further studies of factors that may regulate AML1-ETO via the 3′UTR should focus on the first 3.7 kb of the 3′UTR. The role of AML1-ETO 3′UTR isoform usage in t(8;21) leukemogenesis is difficult to determine because wild type ETO expression is undetectable in healthy HSPCs. However, future studies to examine how modulation of AML1-ETO 3′UTR usage affects AML1-ETO expression would be of interest and may lead to novel therapeutic strategies.
Our study further analyzed the contribution of different regulatory regions within the AML1-ETO 3′UTR to AML1-ETO expression. Our luciferase assay experiments using progressive AML1-ETO 3′UTR fragments showed up to 7-fold differences in luciferase activity between the fragments, with the lowest expression in fragments near the 3’ end of the 3.7 kb 3'UTR. These data suggest that the expression of AML1-ETO is controlled by negatively acting regulatory elements near the 3’ end of the 3.7 kb 3’UTR isoform. Strikingly, both the Kasumi-1 and SKNO-1 t(8;21) AML cell lines shared this trend, suggesting that these regulatory elements may be important in controlling the dosage of AML1-ETO for t(8;21) leukemic maintenance. Our DICER knockdown experiments showed a ~ 3-fold increase in AML1-ETO levels, which is consistent with other reported miRNA regulated genes [62, 63] and may be even greater than observed because DICER knockdown was incomplete. These results suggest that miRNAs are involved in the negative regulation of AML1-ETO, consistent with previous reports [52,53,54]. We further show that this miRNA-mediated regulation affects the AML1-ETO 3’UTR fragment between 2.8 and 3.4 kb. It is likely that additional trans-factors, such as RNA binding proteins, are also involved in the post-transcriptional regulation of AML1-ETO. Our luciferase reporter analysis of the AML1-ETO 3′UTR may be useful in further studies of AML1-ETO post-transcriptional regulation along this line.
Our study identifies let-7b as a direct regulator of the AML1-ETO oncogene. The let-7 family of miRNAs are well described tumor suppressors and are known to target several oncogenes including RAS, MYC and HMGA2 [61, 64,65,66]. Indeed, we found that overall and disease-free survival was significantly higher in let-7b high expression patients within the favorable-risk AML category, of which t(8;21) AML belongs, in the TCGA adult AML dataset [61]. We further demonstrate that transient expression of let-7b inhibits t(8;21) AML proliferation, rescues AML1-ETO target expression, and promotes differentiation. Although additional let-7b targets likely also contribute, these results agree with previous studies showing that directly silencing AML1-ETO with siRNA or shRNA releases the AML1-ETO mediated differentiation block of t(8;21) AML cells as evidenced by down-regulation of the hematopoietic stem cell marker CD34, and up-regulation of more mature myeloid markers such as CD38, CD33, and CD13 [28,29,30, 32, 33]. Identifying endogenous miRNAs that regulate AML1-ETO, such as let-7b, offers a potential method to silence AML1-ETO beyond the direct delivery of exogenous synthetic siRNAs. For example, pre-clinical strategies have been used to upregulate let-7 miRNA expression using small molecule inhibitors against the endogenous protein inhibitors of let-7 biogenesis, such as LIN28A/LIN28B [67,68,69], TUT4 [70, 71], or ADAR1 [72].
Conclusions
Collectively, our study revealed the 3’UTR isoform usage of AML1-ETO in t(8;21) AML, examined regulatory regions throughout the AML1-ETO 3′UTR, and identified let-7b as a novel regulator of AML1-ETO. We further demonstrate that this regulation can inhibit t(8;21) AML cell line proliferation, partially reverse the AML1-ETO mediated differentiation block, and affect AML1-ETO transcriptional targets. Consequently, the post-transcriptional regulation of AML1-ETO through let-7b contributes to the leukemic phenotype of t(8;21) AML and may be important for t(8;21) leukemogenesis and maintenance.
Availability of data and materials
RNA-sequencing (RNA-seq) data of patient samples, healthy, Kasumi-1, and SKNO-1 RNA-seq data was deposited at the European Nucleotide Archive, study accession number: PRJEB42786.
Abbreviations
- AML:
-
Acute myeloid leukemia
- HSPC:
-
Hematopoietic stem and progenitor cell
- miRNA:
-
MicroRNA
- RHD:
-
Runt homology domain
- UTR:
-
Untranslated region
References
Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–47.
Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100(13):4325–36.
Suvajdzic N, Novkovic A, Djunic I, Virijevic M, Colovic N, Vidovic A, et al. Prognostic Factors and Outcome of Core Binding Factor Acute Myeloid Leukemia Patients. Haematologica. 2012;97:274–5.
Bhatt VR, Kantarjian H, Cortes JE, Ravandi F, Borthakur G. Therapy of core binding factor acute myeloid leukemia: incremental improvements toward better long-term results. Clin Lymphoma Myeloma Leuk. 2013;13(2):153–8.
Kurosawa S, Miyawaki S, Yamaguchi T, Kanamori H, Sakura T, Moriuchi Y, et al. Prognosis of patients with core binding factor acute myeloid leukemia after first relapse. Haematologica. 2013;98(10):1525–31.
Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark Ed). 2012;17:1120–39.
Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070–82.
Ibanez V, Sharma A, Buonamici S, Verma A, Kalakonda S, Wang JX, et al. AML1-ETO decreases ETO-2 (MTG16) interactions with nuclear receptor corepressor, an effect that impairs granulocyte differentiation. Cancer Res. 2004;64(13):4547–54.
Lindberg SR, Olsson A, Persson AM, Olsson I. The Leukemia-associated ETO homologues are differently expressed during hematopoietic differentiation. Exp Hematol. 2005;33(2):189–98.
Migasa AA, Mishkova OA, Ramanouskaya TV, Ilyushonak IM, Aleinikova OV, Grinev VV. RUNX1T1/MTG8/ETO gene expression status in human t(8;21)(q22;q22)-positive acute myeloid leukemia cells. Leukemia Res. 2014;38(9):1102–10.
Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112(4):1392–401.
Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18(12):7185–91.
Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18(12):7176–84.
Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001;21(19):6470–83.
Wang L, Gural A, Sun XJ, Zhao XY, Perna F, Huang G, et al. The Leukemogenicity of AML1-ETO Is Dependent on Site-Specific Lysine Acetylation. Science. 2011;333(6043):765–9.
Shia WJ, Okumura AJ, Yan M, Sarkeshik A, Lo MC, Matsuura S, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119(21):4953–62.
Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBP alpha in t(8;21) myeloid leukemia. Nat Med. 2001;7(4):444–51.
Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101(1):270–7.
Klampfer L, Zhang J, Zelenetz AO, Uchida H, Nimer SD. The AML1/ETO fusion protein activates transcription of BCL-2. P Natl Acad Sci USA. 1996;93(24):14059–64.
Zhang JS, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305(5688):1286–9.
Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, et al. Identification of a Dynamic Core Transcriptional Network in t(8;21) AML that Regulates Differentiation Block and Self-Renewal. Cell Rep. 2014;8(6):1974–88.
DeKelver RC, Lewin B, Lam K, Komeno Y, Yan M, Rundle C, et al. Cooperation between RUNX1-ETO9a and novel transcriptional partner KLF6 in upregulation of Alox5 in acute myeloid leukemia. PLoS Genet. 2013;9(10):e1003765.
Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12(8):945–9.
Burel SA, Harakawa N, Zhou LM, Pabst T, Tenen DG, Zhang DE. Dichotomy of AML1-ETO functions: Growth arrest versus block of differentiation. Mol Cell Biol. 2001;21(16):5577–90.
Tonks A, Tonks AJ, Pearn L, Pearce L, Hoy T, Couzens S, et al. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia. 2004;18(7):1238–45.
Yuan YZ, Zhou LM, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. P Natl Acad Sci USA. 2001;98(18):10398–403.
Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1(1):63–74.
Heidenreich O, Krauter J, Riehle H, Hadwiger P, John M, Heil G, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101(8):3157–63.
Martinez N, Drescher B, Riehle H, Cullmann C, Vornlocher HP, Ganser A, et al. The oncogenic fusion protein RUNX1-CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004;4:44.
Dunne J, Cullmann C, Ritter M, Soria NM, Drescher B, Debernardi S, et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006;25(45):6067–78.
Martinez Soria N, Tussiwand R, Ziegler P, Manz MG, Heidenreich O. Transient depletion of RUNX1/RUNX1T1 by RNA interference delays tumour formation in vivo. Leukemia. 2009;23(1):188–90.
Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26(8):1829–41.
Spirin PV, Lebedev TD, Orlova NN, Gornostaeva AS, Prokofjeva MM, Nikitenko NA, et al. Silencing AML1-ETO gene expression leads to simultaneous activation of both pro-apoptotic and proliferation signaling. Leukemia. 2014;28(11):2222–8.
Link KA, Lin S, Shrestha M, Bowman M, Wunderlich M, Bloomfield CD, et al. Supraphysiologic levels of the AML1-ETO isoform AE9a are essential for transformation. Proc Natl Acad Sci U S A. 2016;113(32):9075–80.
Mayr C. Regulation by 3’-Untranslated Regions. Annu Rev Genet. 2017;51:171–94.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
Lin SB, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–33.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22.
Wallace JA, O’Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130(11):1290–301.
Nishi M, Eguchi-Ishimae M, Wu Z, Gao W, Iwabuki H, Kawakami S, et al. Suppression of the let-7b microRNA pathway by DNA hypermethylation in infant acute lymphoblastic leukemia with MLL gene rearrangements. Leukemia. 2013;27(2):389–97.
Xu WQ, Huang YM, Xiao HF. [Expression Analysis and Epigenetics of MicroRNA let-7b in Acute Lymphoblastic Leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23(6):1535–41.
Balzeau J, Menezes MR, Cao SY, Hagan JP. The LIN28/let-7 Pathway in Cancer. Front Genet. 2017;8:1–16.
Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest. 2013;123(6):2395–407.
Dai CW, Bai QW, Zhang GS, Cao YX, Shen JK, Pei MF, et al. MicroRNA let-7f is down-regulated in patients with refractory acute myeloid leukemia and is involved in chemotherapy resistance of adriamycin-resistant leukemic cells. Leuk Lymphoma. 2014;55(7):1645–8.
Huang Y, Hong X, Hu J, Lu Q. Targeted regulation of MiR-98 on E2F1 increases chemosensitivity of leukemia cells K562/A02. Onco Targets Ther. 2017;10:3233–9.
Zhou H, Li Y, Liu B, Shan Y, Li Y, Zhao L, et al. Downregulation of miR-224 and let-7 i contribute to cell survival and chemoresistance in chronic myeloid leukemia cells by regulating ST3GAL IV expression. Gene. 2017;626:106–18.
Cao YX, Wen F, Luo ZY, Long XX, Luo C, Liao P, et al. Downregulation of microRNA let-7f mediated the Adriamycin resistance in leukemia cell line. J Cell Biochem. 2019.
Pelosi A, Careccia S, Lulli V, Romania P, Marziali G, Testa U, et al. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2013;32(31):3648–54.
Wright C, Shin JH, Rajpurohit A, Deep-Soboslay A, Collado-Torres L, Brandon NJ, et al. Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiat. 2017;7.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8.
Li YH, Gao L, Luo XF, Wang LL, Gao XN, Wang W, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509.
Fu L, Shi J, Liu A, Zhou L, Jiang M, Fu H, et al. A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int J Cancer. 2017;140(3):653–61.
Zaidi SK, Perez AW, White ES, Lian JB, Stein JL, Stein GS. An AML1-ETO/miR-29b-1 regulatory circuit modulates phenotypic properties of acute myeloid leukemia cells. Oncotarget. 2017;8(25):39994–40005.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4.
Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson AG, et al. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. New Engl J Med. 2013;368(22):2059–74.
Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–8.
Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6.
Stoner SA, Liu KTH, Andrews ET, Liu MD, Arimoto KI, Yan M, et al. The RUNX1-ETO target gene RASSF2 suppresses t(8;21) AML development and regulates Rac GTPase signaling. Blood Cancer Journal. 2020;10(2).
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom N, Kelnar K, Ovcharenko D, et al. The let-7 MicroRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–22.
Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18(5):549–57.
Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34(17):4801–15.
Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–73.
Sampson VB, Rong NH, Han J, Yang QY, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67(20):9762–70.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120(5):635–47.
Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Gene Dev. 2007;21(9):1025–30.
Roos M, Pradere U, Ngondo RP, Behera A, Allegrini S, Civenni G, et al. A Small-Molecule Inhibitor of Lin28. Acs Chem Biol. 2016;11(10):2773–81.
Lim D, Byun WG, Park SB. Restoring Let-7 microRNA Biogenesis Using a Small-Molecule Inhibitor of the Protein-RNA Interaction. Acs Med Chem Lett. 2018;9(12):1181–5.
Wang LF, Rowe RG, Jaimes A, Yu CX, Nam Y, Pearson DS, et al. Small-Molecule Inhibitors Disrupt let-7 Oligouridylation and Release the Selective Blockade of let-7 Processing by LIN28. Cell Rep. 2018;23(10):3091–101.
Lin SB, Gregory RI. Identification of small molecule inhibitors of Zcchc11 TUTase activity. Rna Biol. 2015;12(8):792–800.
Yu CX, Wang LF, Rowe RG, Han A, Ji WY, McMahon C, et al. A nanobody targeting the LIN28:let-7 interaction fragment of TUT4 blocks uridylation of let-7. P Natl Acad Sci USA. 2020;117(9):4653–63.
Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell. 2016;19(2):177–91.
Acknowledgements
The authors thank the UC San Diego Institute for Genomic Medicine (IGM) and their staff for sequencing the RNA-seq libraries.
Funding
This work was supported by National Institutes of Health, National Cancer Institute grants R01CA104509 (D.E.Z), F31CA228324 (D.T.J), and T32CA009523 (D.T.J).
Author information
Authors and Affiliations
Contributions
Contribution: DTJ and DEZ conceived of the study and designed the experimental plan. DTJ performed the in-vitro biological experiments and interpretation. JHZ and EDB collected, annotated, and provided clinical samples for the study. AGD prepared clinical samples for RNA-sequencing. DTJ performed the bioinformatics and other data analyses. DTJ, AGD, and DEZ prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Patient samples were obtained at UC San Diego Health with written consent, following ethics approval and consent in accordance with the university-approved Institutional Review Board protocol.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary material and methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Johnson, D.T., Davis, A.G., Zhou, JH. et al. MicroRNA let-7b downregulates AML1-ETO oncogene expression in t(8;21) AML by targeting its 3′UTR. Exp Hematol Oncol 10, 8 (2021). https://doi.org/10.1186/s40164-021-00204-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40164-021-00204-7