Abstract
Background
Extended-spectrum beta-lactamase (ESBL)-producing Gram-negative bacteria have become a serious threat to global health. Their rapid spread is associated with high mortality due to ineffective antibiotic treatment. To date a regular surveillance of multidrug-resistant (MDR) pathogens in Ethiopia is not established. For this report, published data regarding ESBL-producing bacteria in different health facilities of Ethiopia were reviewed.
Methods
This study collates data from published information on the rates and clinical implications of infection with ESBL-producing Gram-negative bacteria in Ethiopia. A systematic literature search was conducted using PubMed, PubMed Central, Medline, Science Direct and Google scholar from October 2018 to March 2019. Eligible studies were identified by applying quality criteria. The pooled proportion of ESBL-producing Gram-negative bacteria was estimated based on a random effect model. The publication bias and the variation in proportion estimates attributed to heterogeneity were assessed.
Results
Fourteen studies with relevant data were included in the review. In total, 1649 Gram-negative bacteria isolated from 5191 clinical samples were included. The pooled proportion estimate of ESBL-producing Gram-negative bacteria was 50% (95% CI: 47.7–52.5%. Data showed a high level of heterogeneity (I2 = 95%, P < 0.01). ESBL rates varied by species; 65.7% (263/400) in Klebsiella spp., 48.4% (90/186) in Salmonella spp., and 47.0% (383/815) in E. coli. ESBL-encoding genes were reported in 81 isolates: 67 isolates harbored the CTX-M-1 group and 14 isolates TEM. The mortality associated with infections by bacteria resistant to third generation cephalosporins has rarely been investigated. However, two studies reported a mortality of 33.3% (1/3) and 100% (11/11).
Conclusions
In this meta-analysis, the pooled prevalence of ESBL-producing pathogens is alarmingly high. Data on mortality rates is scarce. This highlights the need for establishing and upgrading clinical microbiology laboratories in Ethiopia for routine antibiotic susceptibility testing and extended surveillance of multidrug resistance.
Similar content being viewed by others
Background
Bacterial production of extended-spectrum β-lactamases (ESBL) significantly reduces the efficacy of the most commonly used beta-lactam antibiotics for the empiric therapy of infections caused by putative Gram-negative bacteria [1]. While ESBL enzymes readily hydrolyse penicillins and cephalosporins, they have a far lower affinity for cephamycins and clavulanic acid [1, 2]. These hydrolytic enzymes are encoded by various gene variants. The major groups are TEM (Temoniera), CTX-M (Cefotaximase-Munich), SHV (Sulfhydryl variable), and OXA (oxacillin), all of which have been used for molecular detection of ESBL genes [3,4,5]. These genes are frequently mobile and located on plasmids and are thus transmitted horizontally [5]. These plasmids often contain mobile elements with resistance genes for additional drug classes such as sulfonamides, aminoglycosides and fluoroquinolones. Thus, bacteria carrying these plasmids are very often multi-drug resistant [6,7,8].
Gram-negative bacteria with the capacity to produce ESBL have become a serious global health problem, especially in resource-limited settings [9]. The rapid increase of ESBL-producing bacteria is associated with high mortality due to ineffective antibiotic treatment [10]. The management of patients with multidrug-resistant (MDR) bacteria requires well-staffed clinical units, reliable microbiology service and regular interaction between professional groups.
Due to limited financial resources and restricted supply chains, carbapenems are often unavailable in healthcare facilities in developing countries. Thus, the effective treatment of infections caused by ESBL-producing bacteria is limited, contributing to a high mortality [11].
In East Africa considerable variance in the prevalence of ESBL-producing Gram-negative bacteria between 13.4 and 89.0% has been described [12,13,14]. Understanding the epidemiology of ESBL at a country level is elemental to reinforce effective prevention and control strategies, but systematic surveillance of MDR pathogens in Ethiopia is non-existent. To assess the magnitude of the problem in Ethiopia, data on ESBL-producing bacteria in different regions of Ethiopia were extracted from existing publications, analyzed and summarized in this systematic review.
Methods
To ensure inclusion of relevant information, the study was conducted based on the guideline of the Preferred Reporting Items for Systematic Reviews and Meta Analyses group checklist [15]. The outcomes of interest were the proportion of ESBL-producing bacteria among Gram-negative isolates from samples obtained from human patients in Ethiopia and the associated mortality.
Study area
The study was conducted in Ethiopia; a country situated in the horn of Africa, covering a land area of 1.04 million km2. With a population of 110,14 million people, Ethiopia is the second most populous nation in Africa following Nigeria [16].
Literature search and eligibility criteria
A systematic literature search was conducted on PubMed, PubMed Central, Medline, Science Direct and Google scholar, which are commonly used medical and biomedical databases in Ethiopia and accessible free of charge through different Ethiopian universities, to identify publications between January 1990 and March 2019 relevant for this review. No chargeable databases or databases without a medical focus were used for the literature search. The search strategy included all articles containing the descriptors. Structured search strategies were developed using the vocabulary terms of each database and targeting the “title” and “abstract” fields. The search was conducted by combining the following medical subject heading terms: “ESBL producing Enterobacteriaceae infections”, “Gram-negative infection associated mortality”, and “Ethiopia”, including all study types and populations. Additional publications were identified in the references of the initially identified articles, including systematic reviews and/or meta-analyses. Citation lists of publications meeting eligibility criteria for this meta-analysis were also reviewed.
In order to identify appropriate publications, the following selection criteria were used: a study had to (i) describe at least one pathogenic bacteria genus among the Gram-negative bacteria, (iii) be conducted on human subjects, (iii) isolate and identify bacteria from clinical specimens, (iv) test isolates for sensitivity to at least amoxicillin plus clavulanic acid and 3rd generation cephalosporins, (v) be conducted in Ethiopia, and (vi) be published in English. Therefore, reviews and publications not focusing on ESBL-producing Gram-negative bacteria isolated from human samples in Ethiopia and/or not subjected to antimicrobial sensitivity testing [17] were excluded. The titles and abstracts of the search results were reviewed and all publications were included that described cross-sectional, prospective, observational, and/or randomized controlled trials.
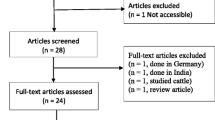
The selection of publications followed three steps. First, titles of articles identified by literature search were checked for relevance of the studied topic. Publications, in which testing for ESBL was not performed, did not describe results from Ethiopia or not from clinical human samples and duplicate publications were discarded. Second, abstracts from selected publications were evaluated for the inclusion criteria. Third, the content of the remaining publications was accessed and evaluated for this review. Details of how eligible studies were included in the data synthesis is indicated in the flow diagram of the study selection process (Fig. 1).
Description of studies
Types of studies
All randomized controlled clinical trials and cross-sectional studies investigating the drug susceptibility/resistance pattern of Gram-negative bacteria from Ethiopia published within the given time frame were included in the review.
Antimicrobial susceptibility testing
Within included studies the double disc synergy test (DDST) was used for phenotype detection of ESBL production showing a typical increase in growth inhibition by ceftazidime and cefotaxime in the zone adjacent to amoxicillin + clavulanic acid [18].
Study populations
The participants of included studies were distributed among all age, sex, and Ethiopian ethnic groups. Overall, 1049 published articles were reviewed. Of these, 14 original articles were included.
Outcome measurements
The main outcome measure was the proportion of ESBL-producing bacteria among Gram-negative isolates evaluated by drug susceptibility testing. Based on this, the isolates defined as resistant to the selected drugs were documented. Patient mortality associated with infections with ESBL-producing Gram-negative bacteria was also recorded.
Data extraction and management
Data was collected from included studies with focus on the following characteristics: type of study, study population, antimicrobial sensitivity testing, and characteristic of the laboratory investigation. For data consistency, two researchers extracted data independently. Whenever there was discordance in the data extracted, consensus was reached by double-checking the article. From eligible studies, if available the following data were extracted: first author, year of publication, study period, study site, study design, population size, sample size, type of sample collection, methods used to test for ESBL, antimicrobial substances used for susceptibility testing, gene encoding for ESBL, number of Gram-negative bacteria isolated and tested for ESBL, the proportion of ESBL-producing bacteria among Gram-negative isolates, and mortality associated with ESBL-producing bacterial infection.
Data synthesis and analysis
The mean proportion of ESBL-producing bacteria was calculated, using the sum of the numbers of ESBL-producing Gram-negative bacteria in all studies considered, divided by the sum of the number of Gram-negative bacteria tested for ESBL. The pooled proportion estimates for ESBL-producing Gram-negative bacteria in the general population and their 95% CI were calculated using the random effects model meta-analysis [19].
Heterogeneity between studies was evaluated through the Cochran’s Q test (reported as p value) and inverse variance index (I2) [20].
For each study, the prevalence with corresponding 95% CI and the overall random effects pooled estimate of all the studies were presented.
Data was analyzed using RStudio Version 1.1.456 –© 2009–2018 (RStudio Inc., Boston, MA, USA). A map showing the number of studies and prevalence of ESBL- producing Gram-negative bacteria in different region of Ethiopia was created, using Quantum GIS software version 2.0.1 (Open Source Geospatial Foundation, Boston, USA).
Results
Distribution of articles describing ESBL in Ethiopia
The initial database search returned 1049 abstracts. Of those, 1018 publications were discarded after reviewing their titles. A further 10 articles either described environmental or veterinary samples or ESBL-producing bacteria were not identified and described in the articles. The full text of 21 articles and an additional three articles identified through review articles were scrutinized for eligibility. Of these, 8 were excluded because ESBL phenotypes or genotypes were not clearly described and 2 publications were identified as systematic reviews. Finally, 14 articles fulfilled eligibility criteria and were subjected to meta-analysis [8, 21,22,23,24,25,26,27,28,29,30,31,32,33,33] (Fig. 1).
In total, the 14 articles reviewed described cross-sectional hospital-based studies. Of those, 6 (43.0%) were published from the cities of Addis Ababa, 5 (36.0%) from Jimma, and 1 (7%) from each of the following: Adama, Bahir Dar, and Harar (Fig. 2). All reviewed articles included patients attending in-patient and/or outpatient departments. A total of 5191 samples from human study participants were analyzed including urine (n = 1273), stool (n = 1679), swabs (n = 266), sputum (n = 294), blood cultures (n = 192) and body fluids (n = 34) (Table 1).
Laboratory methods used to detect ESBL-producing bacteria
Eleven (78.6%) of the reviewed articles used the double disk synergy test (DDST) alone, while the remaining 3 (21.4%) articles used both DDST and polymerase chain reaction (PCR)-based molecular methods to investigate the proportion of ESBL-producing bacteria.
Selected articles were published from 2005 to 2019. Inclusion of study participants ranged from 2003 to 2017. All data in the included publications were obtained from tertiary hospitals. The highest proportions of ESBL-producing bacteria were reported from the capital city, Addis Ababa (57%) and Bahir Dar, the capital of Amhara Region in the northwestern part of Ethiopia (57%). The lowest proportion was reported from Adama in Oromia Region / central Ethiopia (25%) (Fig. 2).
As presented in the forest plot (Fig. 3) the pooled proportion of ESBL-producing Gram-negative bacterial isolates from human samples in Ethiopia was 50% (95% CI: 0.48–0.52). Random model methods showed a high level of heterogeneity (I2 = 95%, p < 0.01). Concerning regional differences, the pooled proportion of ESBL-producing Gram-negative bacteria in Addis Ababa was 56.5% (95% CI, 0.53–0.60; I2 = 95.2% and p < 0.0001) compared to 41% (95% CI, 0.36–0.46; I2 = 84.6% and p < 0.0001) in the city of Jimma in the southwestern part of Ethiopia.
The studies included were conducted in different clinical settings, study periods, with differing clinical samples and study populations. This might be an influencing factor for the heterogeneity of the results.
In total, 1649 Gram-negative bacteria isolated from 5191 clinical samples were included. The pooled rate of ESBL-producing Gram-negatives was determined to be 50.1% (95% CI: 47.7 –52.5%). Among different species, ESBL production rates were 65.7% (n = 263) for Klebsiella spp., 62.2% (n = 33) for Enterobacter spp., 48.4% (n = 90) for Salmonella spp., 47.0% (n = 383) for E. coli, 46.8% (n = 22) for Citrobacter spp., 43.8% (n = 7) for Providencia spp., 28.3% (n = 15) for Proteus spp., 17.4% (n = 4) for Pseudomonas aeruginosa, 9.4% (n = 3) for Acinetobacter spp., and 20.8% (n = 5) for other Gram-negative bacteria, respectively (Table 2).
Molecular epidemiology of ESBL in Ethiopia
Out of 14 reviewed articles, only three (21.4%) studies, one from Addis Ababa University Black Lion Hospital and two from Jimma University Specialized Hospital, reported on the underlying resistance genes for the detection of ESBL resistance [8, 23, 28]. A total of 81 isolates were molecular-biologically analyzed for genetic resistance mechanism. All of those were positive for ESBL-encoding genes: 82.7% (67/81) carried CTX-M-1 group and 17.3% (14/81) TEM. All CTX-M-1 group ESBL genes reported from included studies were confirmed to be CTX-M-15 genes [8, 23]. Additional genes detected were: one isolate carrying a blaOXA gene from Addis Ababa, two blaCTX of the CTX-M-9 group [23] and three blaSHV genes only expressed in Enterobacter cloacae isolated in Jimma [8].
Outcome of patients infected with ESBL-producing bacteria
Only two of the analyzed studies reported the mortality rate associated with infections by ESBL-producing pathogens or resistance to 3rd generation cephalosporins. The pooled mortality within these publications was 86% (12/14) [28, 29]. One study from Black Lion Teaching Hospital in Addis Ababa reported a high mortality among patients with Gram-negative bacterial blood stream infection, particularly among patients infected with multi drug resistant bacteria. Twelve out of 20 patients with Gram-negative sepsis died in the hospital. Of those, 91.7% (11/12) were infected with ESBL-producing bacteria [29].
Discussion
Infections caused by ESBL-producing Gram-negative bacteria are increasing at an alarming rate and have become a serious public health threat worldwide. Summarized data of the burden of ESBL-associated antibiotic resistance is limited in Africa. To our knowledge, this is the first systematic review of data from Ethiopia concerning ESBL-producing bacteria from clinical specimens.
In this study, the pooled prevalence of ESBL-producing Gram-negative bacteria was 50.1%. This prevalence lies in the middle of a wide range between 13.4 to 89.0% reported in other studies from East Africa [13, 14]. Among the different species identified and investigated in the studies, Klebsiella spp. were the most frequent ESBL-producing Gram-negative bacteria followed by Enterobacter spp., Salmonella spp., E. coli and Citrobacter spp. (Table 2). Similar findings have been reported from Uganda, with ESBL-rates of 52% for Klebsiella spp. and 44% for E. coli [12]. One recent study from northern Ethiopia also confirmed the highest rate of ESBL-production among Gram-negative isolates from human samples in K. pneumoniae [34].
The most common ESBL-producing non-fermenting Gram-negative rods (NFGN) are Pseudomonas aeruginosa and Acinetobacter spp. [35]. In this review, we identified a relatively low ESBL rate among NFGN bacteria compared with the Enterobacteriales of 17.4 and 9.4% for Pseudomonas aeruginosa and Acinetobacter spp., respectively (Table 2). One study from Jimma, in southwest Ethiopia described a high prevalence of Acinetobacter spp., during screening for possible ESBL-production using 3rd generation cephalosporins alone. However, genotyping results confirmed a low proportion of ESBL production in Acinetobacter spp. at the study site [8]. This shows that genotypic screening for MDR in NFGN might not be beneficial. In this case, the functional and more general approach of phenotypic resistance testing seems superior.
Molecular ESBL gene detection is not commonly practiced in Ethiopia. From all studies included in this review, gene detection was performed for 81 strains out of the 825 ESBL-producing Gram-negative bacteria determined by DDST. The CTX-M-1 group ESBL genes were most frequently detected, with all of these identified as CTX-M-15, followed by blaTEM. Reports from India showed similar results, in which the prevalence of blaCTX-M was highest (82.5%), followed by blaTEM (67.5%) and blaSHV (57.5%) among the ESBL-genes identified from clinical Gram-negative isolates [17]. In the studies included in this review, blaSHV was only detected in three Enterobacter cloacae isolates. Only a single isolate carrying a blaOXA gene from Addis Ababa [24] and two isolates expressing CTX-M-9 group genes from Jimma [8] were described. More studies are needed to establish a comprehensive overview of the distribution of ESBL genes in Ethiopia.
Isolates of Acinetobacter baumannii carrying the carbapenem resistance gene NDM-1 (New Delhi metallo-beta-lactamase) were reported from Jimma [35]. In East Africa, the NDM-1 gene was first detected from isolated Acinetobacter baumannii in Kenya [36]. However, the current data on the prevalence of carbapenem resistance in East Africa ranges from 1% in the Democratic Republic of Congo to 35% in Tanzania [37], although no systematic surveillance is conducted in these countries and thus, as in Ethiopia, the true prevalence is unknown.
Pooled data currently available suggest that infections caused by ESBL-producing bacteria are leading to prolonged hospital stays and are associated with an increased risk of mortality [11, 38]. Even though available data from Ethiopia is very limited concerning the outcome of patients infected with ESBL-producing bacteria, one study from Addis Ababa describes a mortality rate of 100% in 11 patients [29]. In patients with sepsis caused by Gram-negative bacteria, mortality is strongly associated with antibiotic resistance [39, 40]. The absence of timely microbiology reports in patients with Gram-negative bacterial sepsis compromises successful antimicrobial treatment and possibilities for antimicrobial stewardship. However, even if pertinent resistance patterns would be available earlier, the frequent unavailability and high prices of effective antibiotics greatly contributes to morbidity and mortality associated with these infections.
As a study conducted in Tanzania indicates, inappropriate antibiotic use and two-week fatality rates were significantly higher among patients with septicemia due to ESBL-producing organisms than among those with infections due to non-ESBL producing bacteria [40]. The 30-day and 90-day survival rates of patients infected by MDR bacteria were also significantly lower than patients infected by non-MDR bacteria (58.8% vs. 75.0% at 30 days and 43% vs. 63%, at 90 days) [41]. In a study published in the UK, mortality due to blood stream infections caused by ESBL-producing E. coli was significantly higher than from non-ESBL E. coli [42]. In countries and regions with high rates of ESBL carrying bacteria, as demonstrated here for Ethiopia, these findings have serious implications for empirical antibiotic prescription practice. In Ethiopia, cephalosporins should be considered ineffective in up to 66% of infections with Klebsiella spp. and 47% of cases of E. coli infections as was reported in 2007 for ESBL-E. coli [42].
In order to guide empiric therapy, continuous surveillance of common resistance patterns of Gram-negative isolates and monitoring of ESBL genes circulating in the population are essential. For this task, clinical microbiology laboratories need to be established, upgraded and maintained. In this review, CTX-M-15 ESBL genes were the most frequently detected ESBL gene. At this time, this gene could be used as target for ESBL screening programs in Enterobacteriales in the country, where high-quality and reliable bacteriological laboratories are still rather rare. In general, the molecular testing for blaCTX-M-1 or blaCTX-M-15 would provide a reliable prediction of overall resistance due to ESBL [43]. In order to reduce the dissemination of antibiotic resistance within the country, infection prevention and control measures and the establishment of antimicrobial stewardship programs should be strengthened.
This review is limited by the fact that available literature does not permit a meta-analysis of adjusted mortality associated with infections caused by ESBL-producing bacteria, as only two of the 14 included studies reported the results of patient outcome. Thus, only crude mortality but neither attributable mortality, nor causality were described. Therefore, this meta-analysis highlights the deficiencies of the existing literature to calculate adverse outcome attributable to ESBL-associated resistance. Because of possible clinical implications it would be interesting to describe the proportion of ESBL-positive Enterobacterales in the different clinical specimen (blood, urine. Etc.). However, it was not possible to list proportions of ESBL-producing isolates in different clinical specimen, because it was not stated respective of clinical samples in most articles included into this study. Since ESBL genotypes were only reported in two studies, the pooled prevalence of infections due to ESBL phenotype in Ethiopia was analyzed.. Clinical Laboratory Standards Institute (CLSI) or European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines were used to interpret AST results by all investigators of studies included in this review.
Conclusion and recommendations
In this meta-analysis, the pooled prevalence of phenotypic ESBL production among Gram-negative isolates from human samples is remarkably high. Despite the scarcity of data, infections caused by ESBL-producing bacteria are very likely resulting in an increased mortality. In resource-limited settings, double-disk synergy test can be implemented for screening of ESBL production. The CTX-M-1 group is the predominantly detected ESBL genotype with all of the detected genes confirmed to be CTX-M-15 genes. The CTX-M-1 group or CTX-M-15 gene could therefore be targeted for rapid genetic ESBL screening in the country, thus providing early essential information for decisions on appropriate and effective treatment. In order to provide physicians with urgently needed guidance for antimicrobial therapies according to detected antimicrobial resistance patterns, establishment, upgrading and maintaining of clinical microbiology laboratories in the country capable of reliable double-disk synergy testing are essential. The possibility of ESBL gene detection for routine surveillance is desirable. National and regional treatment guidelines should be based upon MDR surveillance to effectively treat patients and prevent the spread of resistance genes in hospitals and communities. Emphasis for production of antibiotic substances in the country or import from abroad should consider recent MDR surveillance data.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Change history
22 August 2020
An amendment to this paper has been published and can be accessed via the original article.
Abbreviations
- AST:
-
Antimicrobial susceptibility testing
- CI:
-
Confidence interval
- CLSI:
-
Clinical Laboratory Standards Institute
- CTX-M:
-
Cefotaximase-Munich
- DDST:
-
Double disc synergy test
- ESBL:
-
Extended spectrum beta-lactamase
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MDR:
-
Multidrug-resistant
- NDM:
-
New Delhi metallo-beta-lactamase
- NFGN:
-
non-fermenting Gram-negative rods
- OXA:
-
Oxacillin
- PCR:
-
polymerase chain reaction
- SHV:
-
Sulfhdryl variabl
- TEM:
-
Temoniera
References
Rawat D, Nair D. Extended-spectrum beta-lactamases in gram negative Bacteria. J Global Infect Dis. 2010;2:263–74.
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86.
Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965;208:239–41.
Tangden T, Cars O, Melhus A, Lowdin E. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother. 2010;54:3564–8.
Ur Rahman S, Ali T, Ali I, Khan NA, Han B, Gao J. The growing genetic and functional diversity of extended Spectrum Beta-lactamases. Biomed Res Int. 2018;2018:9519718.
Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res Notes. 2010;3:348.
Wang J, Stephan R, Karczmarczyk M, Yan Q, Hachler H, Fanning S. Molecular characterization of Bla ESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front Microbiol. 2013;4:188.
Zeynudin A, Pritsch M, Schubert S, Messerer M, Liegl G, Hoelscher M, Belachew T, Wieser A. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum beta-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect Dis. 2018;18:524.
Vernet G, Mary C, Altmann DM, Doumbo O, Morpeth S, Bhutta ZA, Klugman KP. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis. 2014;20:434–41.
Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D. Ready for a world without antibiotics? The Pensieres antibiotic resistance call to action. Antimicrob Resist Infect Control. 2012;1:11.
Hawkey PM, Warren RE, Livermore DM, McNulty CAM, Enoch DA, Otter JA, Wilson APR. Treatment of infections caused by multidrug-resistant gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association joint working party. J Antimicrob Chemother. 2018;73:iii2–iii78.
Andrew B, Kagirita A, Bazira J. Prevalence of extended-Spectrum Beta-lactamases-producing microorganisms in patients admitted at KRRH, southwestern Uganda. Int J Microbiol. 2017;2017:3183076.
Ampaire L, Nduhura E, Wewedru I. Phenotypic prevalence of extended spectrum beta-lactamases among enterobacteriaceae isolated at Mulago National Referral Hospital: Uganda. BMC Res Notes. 2017;10:448.
Sonda T, Kumburu H, van Zwetselaar M, Alifrangis M, Lund O, Kibiki G, Aarestrup FM. Meta-analysis of proportion estimates of extended-Spectrum-Beta-lactamase-producing Enterobacteriaceae in East Africa hospitals. Antimicrob Resist Infect Control. 2016;5:18.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
World Population Prospects. 2019. Ethiopia Population. http://worldpopulationreview.com/countries/ethiopia-population/.
Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of extended Spectrum beta-lactamase (ESBL) producing gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res. 2013;7:2173–7.
Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008;14(Suppl 1):90–103.
Hedges LV, Vevea JL. Fixed- and random-effects models in meta-analysis. Psychol Methods. 1998;3:486–504.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011;5:23–33.
Desta K, Woldeamanuel Y, Azazh A, Mohammod H, Desalegn D, Shimelis D, Gulilat D, Lamisso B, Makonnen E, Worku A, Mannerqvist K, Struwe J, Aspevall O, Aklillu E. High gastrointestinal colonization rate with extended-Spectrum beta-lactamase-producing Enterobacteriaceae in hospitalized patients: emergence of Carbapenemase-Producing K. pneumoniae in Ethiopia. PLoS One. 2016;11:e0161685.
Eguale T, Asrat D, Alemayehu H, Nana I, Gebreyes WA, Gunn JS, Engidawork E. Phenotypic and genotypic characterization of temporally related nontyphoidal Salmonella strains isolated from humans and food animals in Central Ethiopia. Zoonoses Public Health. 2018;65:766–76.
Gashaw M, Berhane M, Bekele S, Kibru G, Teshager L, Yilma Y, Ahmed Y, Fentahun N, Assefa H, Wieser A, Gudina EK, Ali S. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: a cross sectional study. Antimicrob Resist Infect Control. 2018;7:138.
Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27–34.
Gashe F, Mulisa E, Mekonnen M, Zeleke G. Antimicrobial resistance profile of different clinical isolates against third-generation Cephalosporins. J Pharm (Cairo). 2018;2018:5070742.
Mulualem Y, Kasa T, Mekonnen Z, Suleman S. Occurrence of extended spectrum beta (b)-lactamases in multi-drug resistant Escherichia coli isolated from a clinical setting in Jimma University specialized hospital, Jimma, Southwest Ethiopia. East Afr J Public Health. 2012;9:58–61.
Pritsch M, Zeynudin A, Messerer M, Baumer S, Liegl G, Schubert S, Loscher T, Hoelscher M, Belachew T, Rachow A, Wieser A. First report on Bla NDM-1-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect Dis. 2017;17:180.
Seboxa T, Amogne W, Abebe W, Tsegaye T, Azazh A, Hailu W, Fufa K, Grude N, Henriksen TH. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;10:e0144944.
Seid J, Asrat D. Occurrence of extended spectrum beta-lactamase enzymes in clinical isolates of Klebsiella species from Harar region, eastern Ethiopia. Acta Trop. 2005;95:143–8.
Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8:39.
Abayneh M, Tesfaw G, Abdissa A. Isolation of extended-Spectrum beta-lactamase- (ESBL-) producing Escherichia coli and Klebsiella pneumoniae from patients with community-onset urinary tract infections in Jimma University specialized hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2018;2018:4846159.
Abera B, Kibret M, Mulu W. Extended-Spectrum beta (beta)-lactamases and Antibiogram in Enterobacteriaceae from clinical and drinking water Sources from Bahir Dar City, Ethiopia. PLoS One. 2016;11:e0166519.
Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, Amare A, Feleke T, Gizachew M, Tiruneh M. High prevalence of extended-spectrum beta-lactamase-producing gram-negative pathogens from patients attending Felege Hiwot comprehensive specialized hospital, Bahir Dar, Amhara region. PLoS One. 2019;14:e0215177.
CAK L, Oizumi KY, Caleffi-Ferracioli KR, RBdL S, RAFd Pád, Cardoso RF, CTA P, VLD S. β-lactamase-producing Gram-negative bacteria in an intensive care unit in southern Brazil. Braz J Pharm Sci. 2017;53(2):e16111.
Revathi G, Siu LK, Lu PL, Huang LY. First report of NDM-1-producing Acinetobacter baumannii in East Africa. Int J Infect Dis. 2013;17:e1255–8.
Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11:629.
Superti SV, Augusti G, Zavascki AP. Risk factors for and mortality of extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli nosocomial bloodstream infections. Rev Inst Med Trop Sao Paulo. 2009;51:211–6.
Zilberberg MD, Nathanson BH, Sulham K, Fan W, Shorr AF. Multidrug resistance, inappropriate empiric therapy, and hospital mortality in Acinetobacter baumannii pneumonia and sepsis. Crit Care. 2016;20:221.
Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DS, Urassa WK, Fataki M, Msangi V, Tellevik MG, Maselle SY, Langeland N. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar Es Salaam, Tanzania. J Clin Microbiol. 2005;43:745–9.
Chaisathaphol T, Chayakulkeeree M. Epidemiology of infections caused by multidrug-resistant gram-negative bacteria in adult hospitalized patients at Siriraj hospital. J Med Assoc Thail. 2014;97(Suppl 3):S35–45.
Melzer M, Petersen I. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J Infect. 2007;55:254–9.
Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci Rep. 2019;9:4224.
Acknowledgments
We are grateful for the support of the Bayer Foundation’s Talents for Afria program for the PhD student of the Hirsch Institute for Tropical Medicine in Asella, Ethiopia.
Authors‘contributions
Tafese Beyene Tufa: Conception and design, acquisition and analysis of the data, drafting of the manuscript. Andre Fuchs: Conception and design, analysis of the data, drafting and revision of the manuscript. Takele Beyene Tufa. Design, acquisition and analysis of the data, drafting of the manuscript. Loraine Stoetter, Drafting and revision of the manuscript. Achim J. Kaasch. Analysis and interpretation of data, drafting of the manuscript. Torsten Feldt: Critical revision of manuscript and approval of final version to be published. Dieter Häussinger: Critical revision of manuscript and approval of final version to be published. Colin R. Mackenzie: Interpretation of data, critical revision of the manuscript and approval of final version to be published.
Funding
There was no influence of the funding organisation on analysis or interpretation of the described data. There review was undertaken as an additional activity parallel to a PhD project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: Author name ‘Torsten Feld’ has been corrected to ‘Torsten Feldt’.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tufa, T.B., Fuchs, A., Tufa, T.B. et al. High rate of extended-spectrum beta-lactamase-producing gram-negative infections and associated mortality in Ethiopia: a systematic review and meta-analysis. Antimicrob Resist Infect Control 9, 128 (2020). https://doi.org/10.1186/s13756-020-00782-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-020-00782-x