Abstract
Despite the threat of Fusarium dieback posed due to ambrosia fungi cultured by ambrosia beetles such as Euwallacea spp., the wood-degradation mechanisms utilized by ambrosia fungi are not fully understood. In this study, we analyzed the 16S rRNA and 18S rRNA genes of the microbial community from the Ficus tree tunnel excavated by Euwallacea interjectus and isolated the cellulose-degrading fungus, Fusarium spp. strain EI, by enrichment culture with carboxymethyl cellulose as the sole carbon source. The cellulolytic enzyme secreted by the fungus was identified and expressed in Pichia pastoris, and its enzymatic properties were characterized. The cellulolytic enzyme, termed FsXEG12A, could hydrolyze carboxymethyl cellulose, microcrystalline cellulose, xyloglucan, lichenan, and glucomannan, indicating that the broad substrate specificity of FsXEG12A could be beneficial for degrading complex wood components such as cellulose, xyloglucan, and galactoglucomannan in angiosperms. Inhibition of FsXEG12A function is, thus, an effective target for Fusarium dieback caused by Euwallacea spp.
Similar content being viewed by others
Introduction
Euwallacea spp. is a genus of ambrosia beetles distributed over Asia into Israel, Central America, and in at least five different locations within the United States. These beetles penetrate wood packaging and plant material (Haack 2006; Kirkendall and Ødegaard 2007; O’Donnell et al. 2015; Ploetz et al. 2016; Wingfield et al. 2010). Ambrosia beetles including Euwallacea spp. carry fungi in specialized structures on their integument called mycangia. Symbiotic fungi (consistent associates) often include only two to three partners per ambrosia beetle species. Euwallacea spp. is a genus of over 40 species within the Xyleborini and is the beetle genus known to cultivate ambrosia fusaria, as their larvae feed on the ectosymbiotic filamentous fungus (Gadd and Loos 1947). Euwallacea spp. and their symbiont ambrosial fusaria largely colonize wood from dead or declining species from at least 48 plant families (Aoki et al. 2019; Danthanarayana 1968; Hulcr et al. 2007). They are also known as destructive pests of several economically important woody plants, including Chinese tea (Camellia sinensis L. Kuntze), avocado (Persea americana Mill.), citrus (Citrus spp.), and cacao (Theobroma cacao L.), where they can cause extensive dieback and even death (Brayford 1987). Fusarium dieback is known to be responsible for serious damage mainly to avocado, box elder (Acer negundo L.), castor bean (Ricinus communis L.), and English oak (Quercus robur L.) (Mendel et al. 2012). Recently, the evolutionary histories of key representatives of the Fusarium and Euwallacea clades were reconstructed (O’Donnell et al. 2015). Fusarium spp., termed AF 1–12, were identified, and the dominant fungal symbiont of Euwallacea interjectus was a specialized ambrosia fungus, Fusarium sp. strain AF-3 (O’Donnell et al. 2015).
The effects of symbiotic fungi on the ambrosia beetle host vary from beneficial to neutral to negative (Klepzigl and Six 2004). Most studies have identified these interactions as obligate mutualisms in which the ambrosia beetles rely on nutritional supplementation from the fungi, and the fungi easily colonize the targeted host trees through the beetle’s transport. The ability of ambrosia fungi to degrade and assimilate wood components (e.g. cellulose and hemicellulose) allows for nutritional supplementation to the ambrosia beetles (De Fine Licht and Biedermann 2012). Few studies have conducted a microbial analysis of ambrosia fungi using the denaturing gradient gel electrophoresis (DGGE) method. Despite the threat posed by ambrosia fungi cultured by ambrosia beetles such as Euwallacea spp., the wood-degradation mechanisms employed by ambrosia fungi are not fully understood.
In this study, we analyzed the 16S rRNA and 18S rRNA genes of the microbial community from the Ficus tree tunnel excavated by Euwallacea interjectus. Furthermore, we isolated a cellulose-degrading fungus, termed Fusarium spp. strain EI, by enrichment culture with carboxymethyl cellulose (CMC) as the sole carbon source. The cellulolytic enzyme secreted by the fungus was also identified, expressed in Pichia pastoris, and its enzymatic properties were characterized.
Materials and methods
Specimens
Logs of Ficus carica attacked and bored by E. interjectus (Fig. 1a–c) were sampled from fig orchards in Tokoname, Aichi, Japan (latitude: 35° 11′; longitude: 136° 54′) on 30 September 2009. Tunnels excavated by E. interjectus (mother beetle) (Fig. 1a) were carefully opened to expose the inner surface of the cavities, on which ambrosia fungi were cultivated as larval food (Fig. 1d). Female adults (daughters) emerging from the logs were reared on semi-artificial diets with a two-layer structure as described by Mizuno and Kajimura (2009) to obtain successive generations (Mizuno and Kajimura 2009).
Chemicals and reagents
CMC, xyloglucan, lichenan, curdlan, laminarin, pustulan, glucomannan, and galactomannan were purchased from Megazyme International (Bray, Ireland). MCC was obtained from Funakoshi (Tokyo, Japan).
Polymerase chain reaction (PCR)-DGGE analysis
The microbial community in the tunnel excavated by E. interjectus was harvested by centrifugation at 8000×g for 5 min. The harvested cells were suspended in saline (150 mM NaCl, 10 mM Tris–HCl; pH 8.0) and centrifuged again at 8000×g for 5 min. The total DNA of the microbial community was extracted using an FTA Elute card (GE Healthcare, Waukesha, WI, USA). The 16S/18S rRNA genes were amplified using universal primers (Table 1), as described previously (May et al. 2001; Muyzer et al. 1993). Each amplification reaction mixture (50 μL) consisted of 1 µL of KOD-Plus-Neo DNA polymerase (Toyobo Co., Ltd.; Osaka, Japan), 5 μL of 5× KOD buffer, 5 μL of dNTPs, 1.5 μL of forward and reverse primer (10 µM), 3 μL of template DNA (100 ng), 3 μL of MgSO4 solution (25 mM), and 30 μL of ddH2O. PCR was implemented as follows: after an initial denaturation at 94 °C for 2 min, 25 cycles at 94 °C for 15 s, 50 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min were performed. The PCR amplicons were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), and the DNA concentration was determined by measuring the absorbance at 260 nm. The electrophoresis of the amplification products was performed in 7% (w/v) polyacrylamide gel (37.5:1 acrylamide:bis-acrylamide) in the presence of a linear denaturing gradient that ranged from 40 to 70% using a DCode Mutation Detection System (Bio-Rad, Hercules, CA, USA). DNA bands were detected using a blue light transilluminator (Thermo Fisher Scientific, MA, USA) and were carefully excised. DNA was extracted from the gel pieces by incubation in 50 µL ddH2O for 24 h at 4 °C and then sequenced.
Isolation and identification of cellulose-degrading microorganisms by enrichment culture technique
The microorganisms from the tunnel excavated (Fig. 1c) by E. interjectus were collected and then cultured at 28 °C in MM liquid medium (10 mM NaNO3, 10 mM KH2PO4, 7 mM KCl, 2 mM MgSO4, 2 mL/L Hutner’s trace metals, 1.5% agar; pH 6.5) containing 1.0% CMC as the sole carbon source. In addition, E. interjectus (Fig. 1a) collected from the excavated tunnels in Ficus trees was sprayed and sterilized with 70% ethanol solution. Then, the heads including the mycangia were cut off and aseptically disrupted with a mortar and pestle. The symbiotic microorganisms were also cultured at 28 °C in MM liquid medium containing CMC. After 5 days of incubation, 1 mL of culture supernatant was added to 50 mL of fresh cellulose medium and then cultured at 28 °C. This operation was repeated four times in total. After enrichment culture using MM liquid medium containing CMC, 10 µL of culture supernatant was applied to MM agar medium containing CMC at 28 °C, and then the cellulose-degrading microorganism was isolated. The 18S rRNA and the elongation factor-1α (EF-1α) genes from the isolated fungus from the excavated tunnel and E. interjectus were sequenced using the primers shown in Table 1. Fusarium spp. strain EI has been deposited as NBRC number 113538 in the International Patent Organism Depository, National Institute of Technology and Evaluation (Tokyo, Japan).
Zymogram analysis of extracellular cellulolytic enzymes
Fusarium spp. strain EI was cultured on MM agar medium containing 1.0% CMC at 37 °C for 4 days, and the conidia were harvested. The conidia (1 × 108) were inoculated and cultured in liquid MM medium containing 1.0% CMC as the sole carbon source at 28 °C for 0–6 days. The culture supernatants were concentrated using an Amicon Ultra filter unit (Merck-Millipore, Billerica, MA, USA) and dialyzed against 50 mM Tris–HCl buffer (pH 8.0). All protein collection steps were performed at 4 °C.
To identify the extracellular cellulolytic enzymes from Fusarium spp. strain EI, zymography was performed. Fusarium spp. strain EI was cultured in liquid MM medium containing 1.0% CMC at 28 °C for 3 days, at which time the extracellular proteins secreted from Fusarium spp. strain EI were concentrated by trichloroacetic acid (TCA) treatment, and TCA precipitates were washed with acetone and dried. The concentrated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymogram gels. Zymogram analysis was performed by co-polymerising 0.5% CMC with 12% polyacrylamide gel. After electrophoresis, the gel was soaked for 1 h in 2.5% (v/v) Triton X-100 to remove the SDS and refold the proteins in the gel. The gel was then thoroughly washed three times in MilliQ water and incubated at 30 °C for 60 min in 50 mM acetate buffer (pH 4.0). The gel was stained with 0.1% (w/v) Congo red solution for 30 min. To detect enzyme activity, the gel was de-stained with 1 M of NaCl until pale-red hydrolysis zones appeared against a red background.
Identification of cellulolytic enzymes from Fusarium spp. strain EI
The protein band with cellulose-degrading activity was excised from the gel, digested with trypsin, and analyzed using matrix assisted laser desorption ionisation-time of flight tandem mass spectrometry (MALDI-TOF/TOF–MS), as previously described (Shimizu et al. 2015). Peptide mass fingerprints and MS/MS spectra were analyzed using the MASCOT search engine (Matrix Science Ltd., London, UK) against the database of all entry proteins, as previously described (Sakai et al. 2017a, b).
Cloning of the GH12 gene from Fusarium spp. strain EI
Fusarium spp. strain EI was cultured in MM medium containing CMC as the sole carbon source for 3 days as described above, and total RNA was prepared by using the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands) per the manufacturer’s instructions. Single-strand cDNA was synthesized from total RNA extracted from disrupted fungal cells, as described previously (Shimizu et al. 2009; Sakai et al. 2018). Complementary DNA fragment encoding the GH12 protein was amplified using the primers shown in Table 1. The PCR product was cloned in pPICZα-A (Novagen, Darmstadt, Germany), and the deduced nucleotide sequence was determined using an automated DNA sequencer (CEQ 2000, Beckman Coulter, Brea, CA, USA).
Preparation of recombinant protein
Complementary DNA fragments encoding the GH12 protein gene were prepared by PCR using the oligonucleotide primer set shown in Table 1. The PCR product was digested by EcoRI and then ligated into pPICZα-A (Novagen, Darmstadt, Germany) that had been digested with the same restriction enzyme. The resulting plasmid was introduced into P. pastoris KM71H (Invitrogen, Carlsbad, CA), and the resulting strain was cultured to produce recombinant GH12 protein, as previously described (Kamijo et al. 2019). The culture supernatant was concentrated using an Amicon Ultra filter unit (Merck-Millipore, Massachusetts, USA). The concentrated protein solution was fractionated on a HiTrap Q column (GE Healthcare, Chicago, USA) using a linear gradient of 0–0.5 M NaCl in 50 mM Tris–HCl (pH 8.0). The elution fraction was concentrated using an Amicon Ultra filter unit (Merck-Millipore, Massachusetts, USA). The purified enzyme was cleaved with Endoglycosidase H (New England Biolabs, Ipswich, MA, USA) to remove N-linked glycans according to the manufacturer’s instructions. Deglycosylated protein was applied to a Superose 6 10/300 GL column (GE Healthcare, Chicago, USA), and recombinant protein was eluted with 50 mM Tris–HCl (pH 8.0) containing 50 mM NaCl and dialyzed against 50 mM Tris–HCl (pH 8.0). All protein purification steps were conducted at 4 °C. Protein concentrations were assayed using the Bradford Protein Assay (Bio-Rad Laboratories, California, USA) with bovine serum albumin as the standard.
Enzyme assays
The glycoside hydrolase activity was assayed in 0.5 mL reaction mixtures containing 50 mM acetate buffer (pH 4.0), 0.2–5% (w/v) substrates, and purified recombinant protein. Reactions were incubated at 50 °C, and GH12 protein was removed from the reaction solution using a Nanosep® Centrifugal Device (Pall Corporation, Port Washington, NY, USA), as described in the instruction manual. Flow-through fractions were boiled at 100 °C for 30 min, and the reducing sugars produced by the recombinant protein were measured using 3,5-dinitrosalicylic acid (DNS), as described previously (Sakai et al. 2017a, b). Standard curves were prepared based on solutions containing various concentrations of glucose. One unit of glycoside hydrolase activity was defined as the amount of enzyme required to produce 1 µmol of reducing sugar (glucose equivalents) per minute.
The reaction products released from polysaccharides were separated on TLC Silica gel 60 plates (Merck-Millipore, Massachusetts, USA) using n-butanol:ethanol:water (10:8:7), visualized by staining with 0.82% (v/v) N-(1-naphthyl) ethylenediamine dihydrochloride and 8.6% (v/v) sulfuric acid in ethanol, and baked at 105 °C for 5 min. The reaction products released from xyloglucan and lichenan were analyzed using MALDI-TOF–MS as previously described (Shimizu et al. 2015).
pH stability and thermostability of purified FsXEG12A
The pH stability and thermostability of the GH12 enzyme (i.e. the FsXEG12A) were determined using 1.0% xyloglucan as a substrate. FsXEG12A (1.0 μM) was incubated in 50 mM glycine–HCl (pH 1.0–3.0), 50 mM sodium acetate (pH 3.0–5.0), 50 mM sodium phosphate (pH 5.0–7.0), and 50 mM Tris–HCl (pH 7.0–10.0) at 4 °C for 24 h, and then the residual xyloglucanase (XEG) activity against 1.0% xyloglucan was measured at the optimal pH of 4.0 at 50 °C for 60 min. Purified FsXEG12A was also incubated at temperatures ranging from 40 to 60 °C for 60 min. Residual FsXEG12A activity was assayed at the optimal pH at 50 °C for 60 min. The amount of reducing sugar produced by FsXEG12A was measured using the DNS method (Sakai et al. 2017a, b).
Accession number(s)
The 16S rRNA gene sequences of the bacteria in the excavated tunnel by E. interjectus were deposited in the GenBank database under Accession numbers LC537448 to LC537462, respectively. The 18S rRNA, EF-1α, and FsXEG12A gene sequences of the fungus isolated from the excavated tunnel by E. interjectus were deposited in the GenBank database under Accession numbers LC534254, LC534255, and LC634256, respectively.
Bioinformatics
Nucleotide and amino acid sequences were aligned using ClustalW (Thompson et al. 1994). Phylogenetic analyses of full-length amino acid sequences were conducted via the neighbor-joining method using MEGA 7 software (Tamura et al. 2007).
Results
DGGE profile of tunnels excavated by E. interjectus
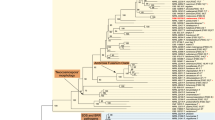
The body length of the adult female E. interjectus was 4–5 mm (Fig. 1a). E. interjectus penetrated the Ficus carica tree (Fig. 1b) and subsequently cultivated ambrosia fungi, which were present in the mycangia or on the exoskeleton. The total DNA was directly extracted from the microbial community in the tunnels excavated by E. interjectus (Fig. 1c, d), and the sequences of 16S rRNA and 18S rRNA genes were amplified using the primer sets shown in Table 1. The DGGE bands of 16S rRNA and 18S rRNA genes were sequenced (Additional file 1: Table S1) and identified using the NCBI database (Table 2). Identified bacteria were categorized into four classes: (1) Brevibacterium sp. (NR_118221.1, 100% identity), Brachybacterium sp. (KX989329.1, 100% identity), Nocardiopsis sp. (MG597576.1, 100% identity), and Streptomyces spp. (KY015030.1, 96%, 99% identity) belonging to Actinobacterium; (2) Facklamia sp. (KU747974.1, 97% identity) and Staphylococcus sp. (KF911124.1, 97% identity) belonging to Firmicutes; (3) Acinetobacter sp. (CP018259.1, 100% identity), Paracoccus sp. (KT345705.1, 98% identity), Bordetella sp. (AF227829.1, 99% identity), and Alcaligenaceae sp. (MF182113.1, 99% identity) belonging to Proteobacterium; and (4) Burkholderia sp. (KM974662.1, 98% identity), Flavobacterium sp. (MF405100.1, 99% identity), and Sphingobacterium sp. (KF911124.1, 97% identity) belonging to Bacteroides (Table 2 and Fig. 2a). The identified fungus belonged to Fusarium spp. (MF522223.1, 99% identity) (Table 2, Fig. 2a and Additional file 1: Fig. S1).
Phylogenetic tree analysis of DGGE profiles of Fusarium isolated from Ficus tree tunnel. a Phylogenetic tree of 16S rRNA and 18S rRNA gene sequences obtained from DGGE analysis. The 16S rRNA gene sequences of the bacteria and the 18S rRNA gene sequence of the fungus in the excavated tunnel by E. interjectus were registered in the GenBank database under Accession numbers LC537448 to LC537462, and LC534254, respectively. The microbial DNA sequences were identified using the NCBI database. b Phylogenetic tree of the partial EF-1α gene sequences of Fusarium spp. isolated from a tunnel in a Ficus tree excavated by E. interjectus, various Fusarium species registered in the JGI database (https://mycocosm.jgi.doe.gov/Fusarium/Fusarium.info.html), and Fusarium sp. AF-3 in the NCBI database. EF-1α genes of KC691533.1 from Fusarium sp. AF-3, KC691536.1 from Fusarium sp. AF-3, 599354 from F. commune MPI-SDFR-AT-0072, 8475 from F. fujikuroi IMI 58289, 9400 from F. graminearum FGSG_08811T0, 17733 from F. oxysporum f. sp. lycopersici 4287, 554906 from F. oxysporum MPI-SDFR-AT-0094, 4767 from F. pseudograminearum CS3096, 485664 from F. solani FSSC 5, 10379 from F. verticillioides 7600, and LC534255 from Fusarium spp. strain EI are shown. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replicates
Isolation and identification of the cellulose-degrading fungus
The microorganisms from the tunnel excavated by E. interjectus (Fig. 1c) were cultured with CMC as the sole carbon source. All 18S rRNA sequences of 10 grown colonies (Additional file 1: Fig. S1) were the same, and sequences of the EF-1α genes, which have been used for DNA typing to resolve all known species within the Ambrosia Fusarium clade (Kasson et al. 2013), were located near that of Fusarium sp. strain AF-3 in the phylogenetic tree (Fig. 2b and Additional file 1: Fig. S2). The fungus isolated in the current study was a species related to Fusarium sp. strain AF-3 and was previously reported to be a symbiotic fungus with E. interjectus (O’Donnell et al. 2015). Therefore, we named this novel strain Fusarium spp. strain EI. The 18S rRNA sequence of Fusarium spp. strain EI corresponded to that of Fusarium spp. as identified above in the DGGE analysis (Table 2 and Additional file 1: Fig. S1). Isolation of fungi from E. interjectus (Fig. 1a) was also performed, and only Fusarium spp. strain EI was isolated using MM liquid medium containing CMC. These results suggested that Fusarium spp. strain EI was a cellulose-degrading symbiotic fungus found in E. interjectus.
Identification of the cellulolytic enzyme produced by Fusarium spp. strain EI
A time-course of cellulolytic activity in the culture supernatant of Fusarium spp. strain EI grown in MM medium containing CMC as the sole carbon source was obtained. Cellulase activity gradually increased, reaching a maximum of 37.4 U/mL at 3 days (Fig. 3a). The extracellular cellulolytic enzymes produced by Fusarium spp. strain EI were analyzed by SDS-PAGE and zymogram gels using CMC as the substrate (Fig. 3b). Four major protein bands were observed on SDS-PAGE, whereas one of the four protein bands was detected when determining cellulolytic activity by zymographic analysis of the culture supernatants (Fig. 3b). A protein band of approximately 25-kDa with cellulolytic activity was identified as EMT67806.1 by peptide mass fingerprinting and MS/MS spectrum analysis using MALDI-TOF/TOF–MS (Fig. 3b and Additional file 1: Table S1). These results indicate that Fusarium spp. strain EI predominantly secretes a cellulolytic enzyme with high similarity to EMT67806.1 in liquid MM medium containing 1.0% CMC.
Time course of cellulase activity in the culture supernatant and zymography analysis of the extracellular proteins. a Cellulolytic activity in culture supernatants from Fusarium spp. strain EI grown with 1.0% CMC for 0–6 days. Data are presented as mean ± standard deviation of three experiments. b Extracellular proteins produced by Fusarium spp. strain EI during a 3-day incubation resolved on SDS-PAGE (left panel) and zymogram gels (right panel). The detected protein bands on SDS-PAGE are indicated by arrows
Cloning, expression, and purification of the 25-kDa protein
For cloning of the full-length DNA encoding the cellulolytic enzyme from Fusarium spp. strain EI, we designed the forward and reverse primers (25-kDa-f and 25-kDa-r) based on the DNA sequence of EMT67806.1 (https://www.uniprot.org/uniprot/N1RQF4#expression). The DNA sequence contained an open reading frame (714 bp) encoding 238 amino acids (Additional file 1: Fig. S3). The N-terminus (amino acids 1–18) was a putative signal peptide sequence. The putative amino acid sequence resembled the sequences of the following annotated proteins: murein transglycosylase from F. oxysporum f. sp. lycopersici 4287 (XP_018258392.1, 100%), probable endoglucanase I precursor from F. proliferatum (CVL13720.1, 97%), murein transglycosylase from F. verticillioides 7600 (XP_018760412.1, 94%), and endoglucanase-1 precursor from F. graminearum PH-1 (XP_011325323.1, 83%). These results showed that the cellulolytic enzyme produced by Fusarium spp. strain EI corresponds to a 25-kDa protein belonging to the glycoside hydrolase family 12 (GH 12). To examine the function of the GH 12 enzyme in detail, it was expressed in P. pastoris (Fig. 4a). For the Pichia expression system, the forward primer without the signal peptide and the reverse primer were designed as 17404-f and 17404-r, respectively (Table 1).
SDS-PAGE of recombinant GH12 enzyme, and TLC and MALDI-TOF–MS analyses of reaction products. a Purified enzyme (1 μg) resolved by SDS-PAGE stained with Coomassie brilliant blue. Lane 1, GH12 enzyme deglycosylated by EndoH; Lane M, protein molecular mass makers. b TLC analysis of soluble products in reaction mixtures with (+) and without (−) enzyme using CMC (lane 1), MCC (lane 2), xyloglucan (lane 3), lichenan (lane 4), curdlan (lane 5), laminarin (lane 6), pustulan (lane 7), glucomannan (lane 8), or galactomannan (lane 9) as the substrate. GH12 enzyme was incubated with 1.0% substrate in 50 mM acetate buffer (pH 3.0) at 50 °C for 60 min. c, d Reaction products from xyloglucan (c) and lichenan (d) were identified using MALDI-TOF–MS
Substrate specificity of the recombinant GH12 enzyme
The substrate specificity of recombinant FsXEG12A was investigated. The GH 12 enzyme hydrolyzed CMC, microcrystalline cellulose (MCC), xyloglucan, lichenan, and glucomannan, while no hydrolase activity was detected towards curdlan, laminarin, pustulan, and galactomannan (Fig. 4b). The reaction products generated by recombinant FsXEG12A from xyloglucan and lichenan were analyzed using MALDI-TOF–MS (Fig. 4c, d). Xyloglucan consists of repeating backbone tetramers of XXXG, XXLG (or XLXG) and XLLG, where G is an unbranched β-d-glucopyranose (β-d-Glcp) residue, X is a β-d-Glcp residue that is decorated by α-1,6-d-xylopyranose (α-d-Xylp), and L is a β-d-Glcp residue with a β-d-galactopyranose (β-d-Galp)-(1 → 2)-α-d-Xylp branch. The main product peaks are consistent with the masses of sodium adducts of XXXG (m/z 1085), XXLG or XLXG (m/z 1247), and XLLG (m/z 1409), and the peaks correspond to sodium adducts of octameric subunits (m/z 2615 and 2778) (Fig. 4c). Similar spectra of the reaction products have been reported previously (Bauer et al. 2005). Lichenan consists of repeating glucose units linked by β-1,3 and β-1,4 glycosidic bonds. The main product peaks detected are consistent with the masses of sodium adducts of dimeric to octameric β-d-Glcp (Fig. 4d). The specific activity of the enzyme towards xyloglucan and lichenan was 1.8- and 1.7-fold of that for CMC, respectively (Additional file 1: Table S2). This indicated that the recombinant GH 12 enzyme specifically recognized and hydrolyzed the β-1,4-glycosidic bond backbone of β-1,4-glucans. The enzyme showed high specific activity toward xyloglucan, suggesting that this enzyme was a XEG (EC 3.2.1.151). Thus, we named the GH12 enzyme as FsXEG12A.
Biochemical characterization of recombinant FsXEG12A
The optimal temperature and pH for FsXEG12A were determined using CMC. The optimal reaction temperature range for FsXEG12A was 20–60 °C (Fig. 5a), whereas the optimal pH was 3.0, with a preferred pH range between 1.0 and 5.0 (Fig. 5b). We determined the kinetic parameters of FsXEG12A using CMC, MCC, xyloglucan, lichenan and glucomannan (Table 3). The catalytic efficiency (kcat/Km) of FsXEG12A was higher toward xyloglucan (1027.0 mL/s/mg) and lichenan (845.2 mL/s/mg) than toward CMC (65.5 mL/s/mg).
Biochemical characterization of FsXEG12A. a Optimal temperature of FsXEG12A. Enzyme reactions proceeded at temperatures ranging from 20 to 100 °C for 60 min. b Optimal pH of FsXEG12A. Enzyme reactions proceeded over a pH range of 1.0–10.0 in 50 mM glycine–HCl (pH 1.0–3.0, filled diamond), 50 mM sodium acetate (pH 3.0–6.0, filled square), 50 mM sodium phosphate (pH 6.0–8.0, filled triangle), and 50 mM Tris–HCl (pH 8.0–10.0, filled circle). c Thermostability of FsXEG12A. Purified FsXEG12A was incubated from 40 to 60 °C for 60 min, and then residual activity against 1.0% CMC was measured after further incubation at 50 °C for 60 min at the optimal pH (pH 3.0). d pH-stability of FsXEG12A. Purified FsXEG12A was incubated in 50 mM glycine–HCl (pH 1.0–3.0, filled diamond), 50 mM sodium acetate (pH 3.0–6.0, filled square), 50 mM sodium phosphate (pH 6.0–8.0, filled triangle) and 50 mM Tris–HCl (pH 8.0–10.0, filled circle) at 4 °C for 24 h. Residual enzyme activity against 1.0% CMC was then measured at the optimal pH (pH 3.0). Data are presented as mean ± standard deviation of three experiments
The thermostability and pH stability of FsXEG12A were investigated using CMC as the substrate. Purified FsXEG12A was incubated at 40 °C, 50 °C, or 60 °C for 60 min (Fig. 5c). After incubation at 40 °C for 60 min, the residual activity of FsXEG12A decreased to 81% of its maximal activity. After incubation at 50 °C for 30 min, the residual activity decreased to 60%. After incubation for 24 h at 4 °C at pH ranging from 1.0 to 10.0, FsXEG12A retained > 60% of its maximal activity (Fig. 5d). These findings indicated that FsXEG12A remains highly stable over a pH range of 1.0 to 10.0.
Discussion
A microbial analysis of 16S rRNA and 18S rRNA genes extracted directly from the microbial community in tunnels excavated by E. interjectus was performed using the DGGE method. Seven gram-positive bacteria, seven gram-negative bacteria, and one fungus were identified (Table 2). Previous studies have shown that E. interjectus is a vector of Ceratocystis ficicola (Kajitani 1996; Morita et al. 2012) and that a wide distribution of C. ficicola in sapwood causes wilt symptoms in trees (Kajii et al. 2013). However, in this study, DGGE analysis identified only Fusarium sp. as a filamentous fungus (Fig. 2a). Fusarium spp. strain EI was isolated from the microorganisms obtained from the excavated tunnels and from E. interjectus. Prospective plant pathogens must overcome the physical barrier presented by the plant cell wall. Plant pathogenic fungi are known for their ability to degrade and assimilate wood components (e.g. cellulose and hemicellulose) and to penetrate the plant cell (Underwood 2012). Our findings identified the E. interjectus-cultured Fusarium spp. strain EI as a cellulose-degrading fungus that produces the XEG FsXEG12A.
Xyloglucan consists of β-1,4-glucan with xylosyl side chains attached to the O-6 position of glycosyl residues, and it associates with cellulose microfibrils through hydrogen bonds, forming a cellulose–xyloglucan network (Damásio et al. 2017; Pauly and Keegstra 2016; Yang et al. 2017). Xyloglucanases (XEGs) are responsible for the hydrolysis of the xyloglucan backbone. Generally, XEG activities are significant towards xyloglucan; however, many XEGs belonging to GH12 and GH74 have low or no activity towards CMC, MCC, lichenan, glucomannan, and xylan (Desmet et al. 2007; Gloster et al. 2007; Sato et al. 2016; Takeda et al. 2013). Highly xyloglucan-specific GH12 XEGs have been isolated from various fungi including Aspergillus niveus (Damásio et al. 2012), Aspergillus terreus (Vitcosque et al. 2016), Aspergillus niger (Master et al. 2008), and Aspergillus cervinus (Rykov et al. 2019). Unlike xyloglucan-specific GH12 XEGs, some fungal GH12 XEGs with broad substrate specificity have also been reported (Segato et al. 2017; Yang et al. 2017). When the specific activity of FsXEG12A towards xyloglucan is set to 100%, its activities towards CMC, MCC, lichenan, and glucomannan are 59%, 21%, 98%, and 45%, respectively (Additional file 1: Table S2). These results indicate that the specific activities of FsXEG12A towards the substrates are at least equal to those of other GH12 XEGs with broad substrate specificity (Segato et al. 2017; Yang et al. 2017). FsXEG12A degrades both xyloglucan and cellulose, resulting in weakening of the cellulose–xyloglucan network. This allows the fungus to infect the plants and utilize the reaction products of cellulose and xyloglucan for fungal growth. Therefore, the broad substrate specificity of FsXEG12A from Fusarium spp. strain EI is beneficial for degrading the complex wood components such as cellulose, xyloglucan, galactoglucomannan, and xylan, which compose 30–40%, 20–25%, 4–8%, and 12–18%, respectively, of the mass of angiosperm plants (Rytioja et al. 2014).
The virulence factors secreted from F. oxysporum have been found to function at different stages of the infection process to induce disease and counteract the plant defence reactions (Roncero et al. 2003). F. oxysporum can produce several toxins such as fusaric acid, beauvericin, enniatin B, bikaverin, moniliformin, fumonisin, and trichothecenes (Irzykowska et al. 2012; Mirocha et al. 1989; Moretti et al. 2002; Son et al. 2008). The potential roles of these toxins in the pathogenic process are not fully understood. Recently, Ma et al. (2015) reported that XEG1 belonging to GH12 not only acts as an XEG but also works as a pathogen-associated molecular pattern (PAMP) in angiosperm plants (for example, soybean and solanaceous species) and that XEG1 triggers cell death (Ma et al. 2015). Transcriptional responses involved in PAMP have been shown to occur in apples, which belong to the order Rosales, as are figs (Puławska et al. 2017). This suggests that the production of GH12 proteins, including FsXEG12, triggers cell death in plants, thereby facilitating infection. FsXEG12 produced by Fusarium spp. strain EI degrades wood components and is critical as a potential virulence factor. Infection with Fusarium spp. could, thus, result in progressive wilting, which often occurs by the blockage of dead vessels through mycelial growth leading to the death of the plant.
For many plant pathogens, including Fusarium species, fungicides have been applied for disease management of the host. Some benzoic acid analogues such as gallic acid, ferulic acid, p-hydroxybenzoic acid, and cuminic acid strongly inhibit F. oxysporum spp. growth (Sun et al. 2017; Wu et al. 2009, 2010a, b), suggesting that these biofungicides could be used for inhibiting the Fusarium dieback mediated by Euwallacea spp. It has also been reported that vanillin, syringaldehyde, trans-cinnamic acid, and hydroxybenzoic acid inhibit cellulose hydrolysis in wet cake (Qin et al. 2016). Therefore, benzoic acid analogues could be used as lead compounds for the design of inhibitors against the cellulase secreted by Fusarium species.
In this study, the cellulose-degrading fungus Fusarium spp. strain EI was isolated from the microbial community cultivated by E. interjectus. Our biochemical characterization of the FsXEG12A enzyme produced by the fungus suggests that the broad substrate specificity of FsXEG12A facilitates the broad species specificity of E. interjectus. Inhibition of FsXEG12A function is, thus, an effective target for reducing Fusarium dieback caused by Euwallacea spp.
Availability of data and materials
Corresponding author could provide all the experimental data on reasonable request.
References
Aoki T, Smith JA, Kasson MT, Freeman S, Geiser DM, Geering ADW, O’Donnell K (2019) Three novel Ambrosia Fusarium clade species producing clavate macroconidia known (F. floridanum and F. obliquiseptatum) or predicted (F. tuaranense) to be farmed by Euwallacea spp. (Coleoptera: Scolytinae) on woody hosts. Mycologia 111:919–935
Bauer S, Vasu P, Mort AJH, Somerville CR (2005) Cloning, expression, and characterization of an oligoxyloglucan reducing end-specific xyloglucanobiohydrolase from Aspergillus nidulans. Carbohydr Res 340:2590–2597
Brayford D (1987) Fusarium bugnicourtii sp. nov., and its relationship to F. tumidum and F. tumidum var. coeruleum. Trans Br Mycol Soc 89:347–351
Damásio ARL, Ribeiro LF, Ribeiro LF, Furtado GP, Segato F, Almeida FBR, Crivellari AC, Buckeridge MS, Souza TA, Murakami MT, Warg RJ, Prade RA, Polizeli ML (2012) Functional characterization and oligomerization of a recombinant xyloglucan-specific endo-β-1,4-glucanase (GH12) from Aspergillus niveus. Biochim Biophys Acta 1824:461–467
Damásio AR, Rubio MV, Gonçalves TA, Persinoti GF, Segato F, Prade RA, Contesini FJ, de Souza AP, Buckeridge MS, Squina FM (2017) Xyloglucan breakdown by endo-xyloglucanase family 74 from Aspergillus fumigatus. Appl Microbiol Biotechnol 101:2893–2903
Danthanarayana W (1968) The distribution and host-range of the shot-hole borer (Xyleborus fornicatus Eich.) of tea. Tea Quart 39:61–69
De Fine Licht HH, Biedermann PH (2012) Patterns of functional enzyme activity in fungus farming ambrosia beetles. Front Zool 9:1–11
Desmet T, Cantaert T, Gualfetti P, Nerinckx W, Gross L, Mitchinson C, Piens K (2007) An investigation of the substrate specificity of the xyloglucanase Cel74A from Hypocrea jecorina. FEBS J 274:356–363
Gadd CH, Loos CA (1947) The ambrosia fungus of Xyleborus fornicatus Eich. Trans Br Mycol Soc 31:13–18
Gloster TM, Ibatullin FM, Macauley K, Eklöf JM, Roberts S, Turkenburg JP, Bjørnvad ME, Jørgensen PL, Danielsen S, Johansen KS, Borchert TV, Wilson KS, Brumer H, Davies GJ (2007) Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem 282:19177–19189
Haack RA (2006) Exotic bark- and wood-boring Coleoptera in the United States: recent establishments and interceptions. Can J For Res 36:269–288
Hulcr J, Mogia M, Isua B, Novotny V (2007) Host specificity of ambrosia and bark beetles (Col., Curculionidae: Scolytinae and Platypodinae) in a New Guinea rain forest. Ecol Entomol 32:762–772
Irzykowska L, Bocianowski J, Waśkiewicz A, Weber Z, Karolewski Z, Goliński P, Kostecki M, Irzykowski W (2012) Genetic variation of Fusarium oxysporum isolates forming fumonisin B-1 and moniliformin. J Appl Genet 53:237–247
Kajii C, Morita T, Jikumaru S, Kajimura H, Yamaoka Y, Kuroda K (2013) Xylem dysfunction in Ficus carica infected with wilt fungus Ceratocystis ficicola and the role of the vector beetle Euwallacea interjectus. IAWA J 34:301–312
Kajitani Y (1996) The possibility of transmission of fig Ceratocystis canker disease by an ambrosia beetle (Xyleborus interjectus Eichhoff). Ann Phytopathol Soc Jpn 62:275 (in Japanese)
Kamijo J, Sakai K, Suzuki H, Suzuki K, Kunitake E, Shimizu M, Kato M (2019) Identification and characterization of a thermostable pectate lyase from Aspergillus luchuensis var. saitoi. Food Chem 276:503–510
Kasson MT, O’Donnell K, Rooney AP, Sink S, Ploetz RC, Ploetz JN, Konkol JL, Carrillo D, Freeman S, Mendel Z, Smith JA, Black AW, Hulcr J, Bateman C, Stefkova K, Campbell PR, Geering AD, Dann EK, Eskalen A, Mohotti K, Short DP, Aoki T, Fenstermacher KA, Davis DD, Geiser DM (2013) An inordinate fondness for Fusarium: phylogenetic diversity of fusaria cultivated by ambrosia beetles in the genus Euwallacea on avocado and other plant hosts. Fungal Genet Biol 56:147–157
Kirkendall LR, Ødegaard F (2007) Ongoing invasions of old-growth tropical forests: establishment of three incestuous beetle species in Central America (Curculionidae, Scolytinae). Zootaxa 1588:53–62
Klepzigl KD, Six DL (2004) Bark beetle-fungal symbiosis: context dependency in complex associations. Symbiosis 37:189–205
Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, Dong S, Zhang Z, Dou D, Zheng X, Tyler BM, Wang Y (2015) A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27:2057–2072
Master ER, Zheng Y, Storms R, Tsang A, Powlowski J (2008) A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem J 411:161–170
May LA, Smiley B, Schmidt MG (2001) Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can J Microbiol 47:829–841
Mendel Z, Protasov M, Sharon A, Zveibil S, Yehuda Ben, O’Donnell K, Rabaglia R, Wysoki M, Freeman S (2012) An Asian ambrosia beetle Euwallacea fornicatus and its novel symbiotic fungus Fusarium sp. pose a serious threat to the Israeli avocado industry. Phytoparasitica 40:235–238
Mirocha CJ, Abbas HK, Kommedahl T, Jarvis BB (1989) Mycotoxin production by Fusarium oxysporum and Fusarium sporotrichioides isolated from Baccharis spp. from Brazil. Appl Environ Microbiol 55:254–255
Mizuno T, Kajimura H (2009) Effects of ingredients and structure of semi-artificial diet on the reproduction of an ambrosia beetle, Xyleborus pfeili (Ratzeburg) (Coleoptera: Curculionidae: Scolytinae). Appl Entomol Zool 44:363–370
Moretti A, Belisario A, Tafuri A, Ritieni A, Corazza L, Logrieco A (2002) Production of beauvericin by different races of Fusarium oxysporum f. sp. melonis, the Fusarium wilt agent of muskmelon. Eur J Plant Pathol 108:661–666
Morita T, Hara H, Mise D, Jikumaru S (2012) A case study of Ceratocystis canker epidemic in relation with Euwallacea interjectus infestation. Ann Rept Kansai Pl Prot 54:29–34 (in Japanese with English summary)
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
O’Donnell K, Sink S, Libeskind-Hadas R, Hulcr J, Kasson MT, Ploetz RC, Konkol JL, Ploetz JN, Carrillo D, Campbell A, Duncan RE, Liyanage PN, Eskalen A, Na F, Geiser DM, Bateman C, Freeman S, Mendel Z, Sharon M, Aoki T, Cossé AA, Rooney AP (2015) Discordant phylogenies suggest repeated host shifts in the Fusarium-Euwallacea ambrosia beetle mutualism. Fungal Genet Biol 82:277–290
Pauly M, Keegstra K (2016) Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67:235–259
Ploetz RC, Hulcr J, Wingfield MJ, de Beer ZW (2016) Destructive tree diseases associated with ambrosia and bark beetle-associated tree diseases: black swan events in tree pathology? Plant Dis 97:856–872
Puławska J, Kałużna M, Warabieda W, Mikiciński A (2017) Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars—susceptible and resistant to fire blight. BMC Genom 18:868
Qin L, Li WC, Liu L, Zhu JQ, Li X, Li BZ, Yuan YJ (2016) Inhibition of lignin-derived phenolic compounds to cellulase. Biotechnol Biofuels 9:1–10
Roncero MIG, Hera C, Ruiz-Rubio M, García-Maceira FI, Madrid MP, Caracuel Z, Calero F, Delgado-Jarana J, Roldán-Rodriguez R, Martínez-Rocha AL (2003) Fusarium as a model for studying virulence in soilborne plant pathogens. Physiol Mol Plant Pathol 62:87–98
Rykov SV, Kornberger P, Herlet J, Tsurin NV, Zorov IN, Zverlov VV, Liebl W, Schwarz WH, Yarotsky SV, Berezina OV (2019) Novel endo-(1,4)-β-glucanase Bgh12A and xyloglucanase Xgh12B from Aspergillus cervinus belong to GH12 subgroup I and II, respectively. Appl Microbiol Biotechnol 103:7553–7566
Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR (2014) Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Biol Rev 78:614–649
Sakai K, Mochizuki M, Yamada M, Shinzawa Y, Minezawa M, Kimoto S, Murata S, Kaneko Y, Ishihara S, Jindou S, Kobayashi T, Kato M, Shimizu M (2017a) Biochemical characterization of thermostable β-1,4-mannanase belonging to the glycoside hydrolase family 134 from Aspergillus oryzae. Appl Microbiol Biotechnol 101:3237–3245
Sakai K, Kojiya S, Kamijo J, Tanaka Y, Tanaka K, Maebayashi M, Oh JS, Ito M, Hori M, Shimizu M, Kato M (2017b) Oxygen-radical pretreatment promotes cellulose degradation by cellulolytic enzymes. Biotechnol Biofuels 10:1–10
Sakai K, Matsuzaki F, Wise L, Sakai Y, Jindou S, Ichinose H, Takaya N, Kato M, Wariishi H, Shimizu M (2018) Biochemical characterization of CYP505D6, a self-sufficient cytochrome P450 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 84:e01091-18
Sato S, Ohta K, Kojima K, Kozeki T, Ohmachi T, Yoshida T (2016) Isolation and characterization of two types of xyloglucanases from a phytopathogenic fungus, Verticillium dahliae. J Appl Glycosci 63:13–18
Segato F, Dias B, Berto GL, de Oliveria DM, De Souza FHM, Citadini AP, Murakami MT, de Lima Damásio AR, Squina FM, Polikarpov I (2017) Cloning, heterologous expression and biochemical characterization of a non-specific endoglucanase family 12 from Aspergillus terreus NIH2624. Biochim Biophys Acta 1865:395–403
Shimizu M, Fujii T, Masuo S, Fujita K, Takaya N (2009) Proteomic analysis of Aspergillus nidulans cultured under hypoxic conditions. Proteomics 9:7–19
Shimizu M, Kaneko Y, Ishihara S, Mochizuki M, Sakai K, Yamada M, Murata S, Itoh E, Yamamoto T, Sugimura Y, Hirano T, Takaya N, Kobayashi T, Kato M (2015) Novel β-1,4-mannanase belonging to a new glycoside hydrolase family in Aspergillus nidulans. J Biol Chem 290:27914–27927
Son SW, Kim HY, Choi GJ, Lim HK, Jang KS, Lee SO, Lee S, Sung ND, Kim JC (2008) Bikaverin and fusaric acid from Fusarium oxysporum show antioomycete activity against Phytophthora infestans. J Appl Microbiol 104:692–698
Sun Y, Wang Y, Han LR, Zhang X, Feng JT (2017) Antifungal activity and action mode of cuminic acid from the seeds of Cuminum cyminum L. against Fusarium oxysporum f. sp. niveum (FON) causing Fusarium wilt on watermelon. Molecules 22:1–13
Takeda T, Nakano Y, Takahashi M, Sakamoto Y, Konno N (2013) Polysaccharide-inducible endoglucanases from Lentinula edodes exhibit a preferential hydrolysis of 1,3-1,4-β-glucan and xyloglucan. J Agric Food Chem 61:7591–7598
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Underwood W (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci 3:1–6
Vitcosque GL, Ribeiro LF, de Lucas RC, da Silva TM, Ribeiro LF, deLima Damasio AR, Farinas CS, Goncalves AZ, Segato F, Buckeridge MS, Jorge JA, Polizeli ML (2016) The functional properties of a xyloglucanase (GH12) of Aspergillus terreus expressed in Aspergillus nidulans may increase performance of biomass degradation. Appl Microbiol Biotechnol 100:9133–9144
Wingfield MJ, Slippers B, Wingfield BD (2010) Novel associations between pathogens, insects and tree species threaten world forests. N Z J For Sci 40:S95–S103
Wu HS, Wang Y, Zhang CY, Bao W, Ling N, Liu DY, Shen QR (2009) Growth of in vitro Fusarium oxysporum f. sp. niveum in chemically defined media amended with gallic acid. Biol Res 42:297–304
Wu HS, Luo J, Raza W, Liu YX, Gu M, Chen G, Hu XF, Wang JH, Mao ZS, Shen QR (2010a) Effect of exogenously added ferulic acid on in vitro Fusarium oxysporum f. sp. niveum. Sci Hortic 124:448–453
Wu HS, Shen S, Han J, Liu Y, Liu S (2010b) The effect in vitro of exogenously applied p-hydroxybenzoic acid on Fusarium oxysporum f. sp. niveum. Phytopathol Mediterr 48:439–446
Yang H, Shi P, Liu Y, Xia W, Wang X, Cao H, Ma R, Luo H, Bai Y, Yao B (2017) Loop 3 of fungal endoglucanases of glycoside hydrolase family 12 modulates catalytic efficiency. Appl Environ Microbiol 83:e03123-16
Acknowledgements
We are grateful to Dr. Tetsuo Kobayashi for helpful discussion. We would like to thank Editage (http://www.editage.jp) for English-language editing.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (21380055 to MS, 25450116 to MK, 17H03831 to HK) and was partially supported by a Grant from the Toyoaki Scholarship Foundation (H30 to MS).
Author information
Authors and Affiliations
Contributions
KS HK, MK, and MS designed the experiments. KS, AY, ST, YK, TS, HS, SJ, YS, and MS performed the experiments and analyzed data. All authors reviewed the results, contributed to the writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Identification of the cellulolytic enzyme produced by Fusarium spp. strain EI. Table S2. Substrate specificity of the purified GH12 enzyme. Figure S1. Nucleotide sequence alignment of 18S rRNA. Figure S2. Nucleotide sequence alignment of EF-1α. Figure S3. Amino acid sequence alignment of GH12 enzymes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sakai, K., Yamaguchi, A., Tsutsumi, S. et al. Characterization of FsXEG12A from the cellulose-degrading ectosymbiotic fungus Fusarium spp. strain EI cultured by the ambrosia beetle. AMB Expr 10, 96 (2020). https://doi.org/10.1186/s13568-020-01030-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-020-01030-6